4X7M
 
 | | Crystal structure of S. aureus TarM G117R mutant in complex with UDP and UDP-GlcNAc | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X6L
 
 | | Crystal structure of S. aureus TarM in complex with UDP | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7P
 
 | | Crystal structure of apo S. aureus TarM | | Descriptor: | SULFATE ION, TarM | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7R
 
 | | Crystal structure of S. aureus TarM G117R mutant in complex with Fondaparinux, alpha-GlcNAc-glycerol and UDP | | Descriptor: | (2S)-2,3-dihydroxypropyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, TarM, ... | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5TZ8
 
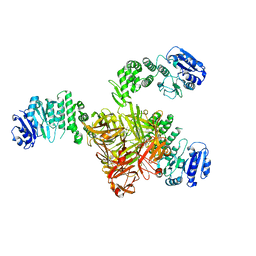 | | Crystal structure of S. aureus TarS | | Descriptor: | Glycosyl transferase | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TZI
 
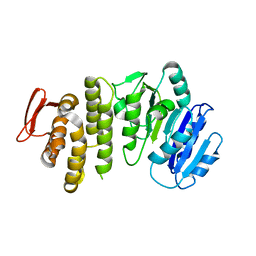 | | Crystal structure of S. aureus TarS 1-349 | | Descriptor: | Glycosyl transferase | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5U02
 
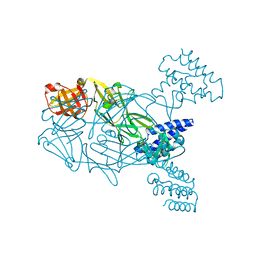 | | Crystal structure of S. aureus TarS 217-571 | | Descriptor: | Glycosyl transferase, IMIDAZOLE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-22 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TZJ
 
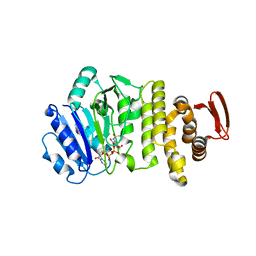 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TZE
 
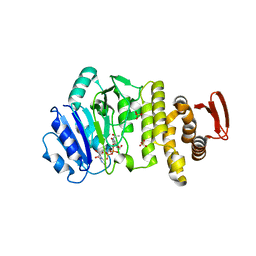 | | Crystal structure of S. aureus TarS in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5TZK
 
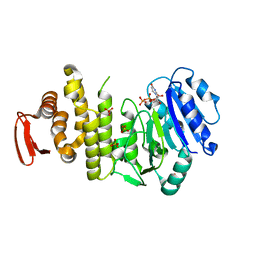 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP | | Descriptor: | Glycosyl transferase, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
8EXP
 
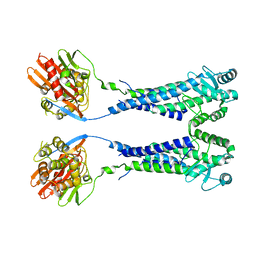 | | Cryo-EM structure of S. aureus BlaR1 with C2 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXS
 
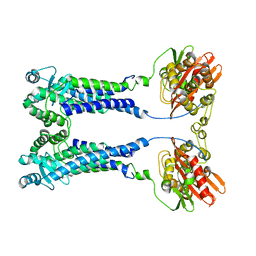 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXR
 
 | | Cryo-EM structure of S. aureus BlaR1 TM and zinc metalloprotease domain | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Beta-lactam sensor/signal transducer BlaR1, PHOSPHATE ION, ... | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXT
 
 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant in complex with ampicillin | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXQ
 
 | | Cryo-EM structure of S. aureus BlaR1 with C1 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
7KMG
 
 | | LY-CoV555 neutralizing antibody against SARS-CoV-2 | | Descriptor: | GLYCEROL, LY-CoV555 Fab heavy chain, LY-CoV555 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7KMH
 
 | | LY-CoV488 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV488 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Boyles, J.S, Dickinson, C.D, Coleman, K.A. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7L3N
 
 | | SARS-CoV 2 Spike Protein bound to LY-CoV555 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV555 Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection.
Biorxiv, 2020
|
|
7KMI
 
 | | LY-CoV481 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV481 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
