1LTS
 
 | |
1LTT
 
 | |
7R2G
 
 | | USP15 D1D2 in catalytically-competent state bound to mitoxantrone stack (isoform 2) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 15, ... | | Authors: | Priyanka, A, Sixma, T.K. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mitoxantrone stacking does not define the active or inactive state of USP15 catalytic domain.
J.Struct.Biol., 214, 2022
|
|
8AG6
 
 | |
9FD2
 
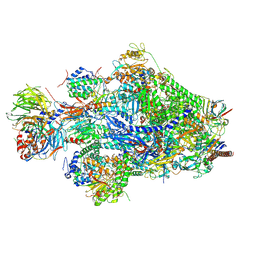 | | Structure of Pol II-TC-NER-STK19 complex | | Descriptor: | DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Lee, S.-H, Sixma, T.K. | | Deposit date: | 2024-05-16 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | STK19 drives transcription-coupled repair by stimulating repair complex stability, RNA Pol II ubiquitylation, and TFIIH recruitment.
Mol.Cell, 84, 2024
|
|
8QH5
 
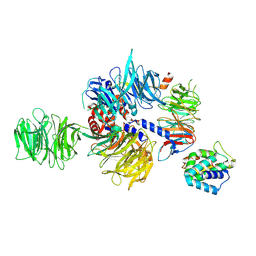 | | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1 | | Descriptor: | DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Lee, S.-H, Sixma, T.K. | | Deposit date: | 2023-09-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1
To Be Published
|
|
5FWI
 
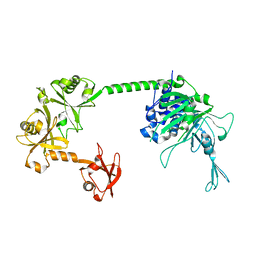 | |
9HNW
 
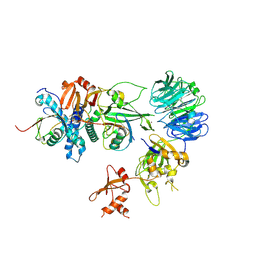 | |
1UV6
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with carbamylcholine | | Descriptor: | 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
1UW6
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
1UX2
 
 | | X-ray structure of acetylcholine binding protein (AChBP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-02-18 | | Release date: | 2004-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
1LTA
 
 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF E. COLI HEAT-LABILE ENTEROTOXIN (LT) WITH BOUND GALACTOSE | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SUBUNIT A, SUBUNIT B, ... | | Authors: | Merritt, E.A, Sixma, T.K, Kalk, K.H, Van Zanten, B.A.M, Hol, W.G.J. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Galactose-binding site in Escherichia coli heat-labile enterotoxin (LT) and cholera toxin (CT).
Mol.Microbiol., 13, 1994
|
|
6YN1
 
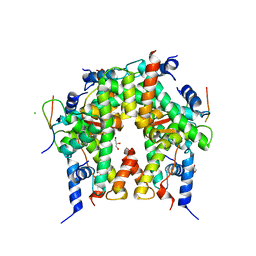 | | Crystal structure of histone chaperone APLF acidic domain bound to the histone H2A-H2B-H3-H4 octamer | | Descriptor: | Aprataxin and PNK-like factor, CHLORIDE ION, GLYCEROL, ... | | Authors: | Corbeski, I, Guo, X, Van Ingen, H, Sixma, T.K. | | Deposit date: | 2020-04-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv, 8, 2022
|
|
6QLY
 
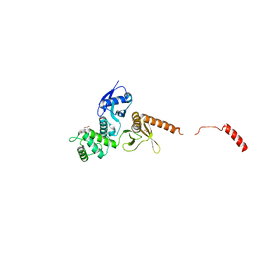 | | IDOL FERM domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | Authors: | Martinelli, L, Sixma, T.K. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
6QML
 
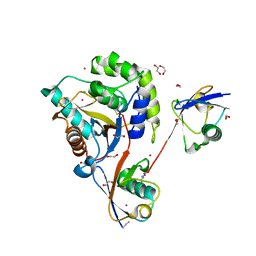 | | UCHL3 in complex with synthetic, K27-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Murachelli, A.G, Sixma, T.K. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | K27-Linked Diubiquitin Inhibits UCHL3 via an Unusual Kinetic Trap.
Cell Chem Biol, 28, 2021
|
|
6I5F
 
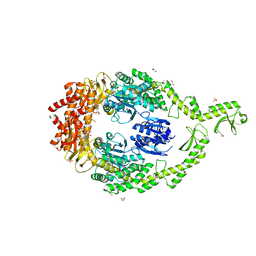 | | Crystal structure of DNA-free E.coli MutS P839E dimer mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, GLYCEROL, ... | | Authors: | Bhairosing-Kok, D, Groothuizen, F.S, Fish, A, Dharadhar, S, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2018-11-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sharp kinking of a coiled-coil in MutS allows DNA binding and release.
Nucleic Acids Res., 47, 2019
|
|
1OH5
 
 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A C:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*CP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH6
 
 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN A:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*AP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP* CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP* CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH8
 
 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN UNPAIRED THYMIDINE | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP* TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*CP*TP*TP*GP*GP*CP* AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH7
 
 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A G:G MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*GP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1LTB
 
 | |
1WB9
 
 | | Crystal Structure of E. coli DNA Mismatch Repair enzyme MutS, E38T mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1WBB
 
 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38A mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1W7A
 
 | | ATP bound MutS | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP* AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP* CP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lamers, M.H, Georgijevic, D, Lebbink, J, Winterwerp, H.H.K, Agianian, B, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
J.Biol.Chem., 279, 2004
|
|
1WBD
 
 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38Q mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
