4L79
 
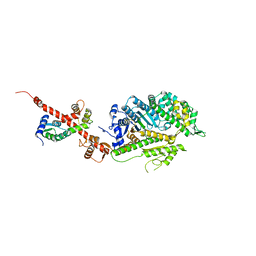 | | Crystal Structure of nucleotide-free Myosin 1b residues 1-728 with bound Calmodulin | | Descriptor: | Calmodulin, MAGNESIUM ION, Unconventional myosin-Ib | | Authors: | Shuman, H, Zwolak, A, Dominguez, R, Ostap, E.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4G1T
 
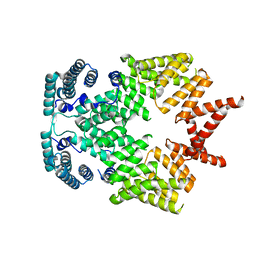 | | Crystal structure of interferon-stimulated gene 54 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 2 | | Authors: | Yang, Z, Liang, H, Zhou, Q, Li, Y, Chen, H, Ye, W, Chen, D, Fleming, J, Shu, H, Liu, Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms.
Cell Res., 22, 2012
|
|
4OFH
 
 | |
4OFE
 
 | |
6U2O
 
 | |
6U6B
 
 | | Structure of human DNA polymerase beta misinserting dAMPNPP opposite the 5'G of the cisplatin Pt-GG intrastrand crosslink with Manganese in the active site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Cisplatin, DNA (5'-D(*CP*CP*CP*AP*CP*GP*GP*CP*CP*CP*AP*TP*CP*AP*CP*C)-3'), ... | | Authors: | Ouzon-Shubeita, H, Vilas, C.K, Lee, S. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Structural insights into the promutagenic bypass of the major cisplatin-induced DNA lesion.
Biochem.J., 477, 2020
|
|
6MXO
 
 | | Structure of HPoleta incorporating dCTP opposite the 3-prime Pt(DACH)-GG | | Descriptor: | (cyclohex-1-ene-1,2-diamine)platinum(2+), 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*CP*GP*GP*CP*TP*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Ouzon-Shubeita, H, Lee, S. | | Deposit date: | 2018-10-31 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis for the bypass of the major oxaliplatin-DNA adducts by human DNA polymerase eta.
Biochem. J., 476, 2019
|
|
5V7X
 
 | | Crystal Structure of Myosin 1b residues 1-728 with bound sulfate and Calmodulin | | Descriptor: | Calmodulin-1, SULFATE ION, Unconventional myosin-Ib | | Authors: | Zwolak, A, Shuman, H, Dominguez, R, Ostap, E.M. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.141 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C1G
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5CHZ
 
 | |
6C1H
 
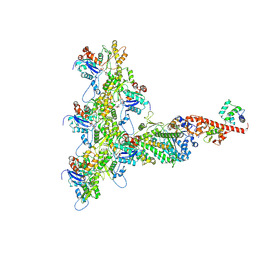 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C1D
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4ETG
 
 | | Crystal Structure of MIF L46G mutant | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Ashrafi, A, Pojer, F, Lashuel, H. | | Deposit date: | 2012-04-24 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterization of molecular determinants of the conformational stability of macrophage migration inhibitory factor: leucine 46 hydrophobic pocket.
Plos One, 7, 2012
|
|
4EVG
 
 | | Crystal Structure of MIF L46A mutant | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Ashrafi, A, Pojer, F, Lashuel, H. | | Deposit date: | 2012-04-26 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of molecular determinants of the conformational stability of macrophage migration inhibitory factor: leucine 46 hydrophobic pocket.
Plos One, 7, 2012
|
|
4EUI
 
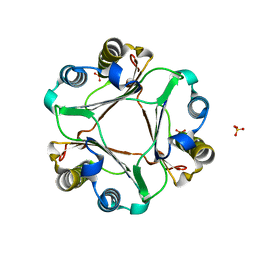 | | Crystal Structure of MIF L46F mutant | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Ashrafi, A, Pojer, F, Lashuel, H. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of molecular determinants of the conformational stability of macrophage migration inhibitory factor: leucine 46 hydrophobic pocket.
Plos One, 7, 2012
|
|
4OFA
 
 | |
4M0M
 
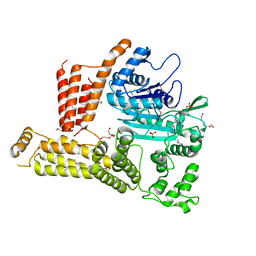 | | The crystal structure of a functionally unknown protein lpg2422 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | PHOSPHATE ION, Putative uncharacterized protein, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Li, H, Clancy, S, Shuman, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | The crystal structure of a functionally unknown protein lpg2422 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
2JM8
 
 | | R21A Spc-SH3 free | | Descriptor: | Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2JMA
 
 | | R21A Spc-SH3:P41 complex | | Descriptor: | P41 peptide, Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2JM9
 
 | | R21A Spc-SH3 bound | | Descriptor: | Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2OIV
 
 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
2OIX
 
 | | Xanthomonas XopD C470A Mutant | | Descriptor: | Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
4FES
 
 | | Structure of OSH4 in complex with cholesterol analogs | | Descriptor: | (3S,8S,9S,10R,13S,14S,17R)-3-hydroxy-10,13-dimethyl-17-[(2S,6S)-6-methyl-3-oxooctan-2-yl]-1,2,3,4,7,8,9,10,11,12,13,14,15,17-tetradecahydro-16H-cyclopenta[a]phenanthren-16-one, Protein KES1 | | Authors: | Koag, M.C, Monzingo, A.F, Cheun, Y, Lee, S. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure of 16,22-diketocholesterol bound to oxysterol-binding protein Osh4.
Steroids, 78, 2013
|
|
4P2H
 
 | |
6VJW
 
 | | Crystal structure of WT hMBD4 complexed with T:G mismatch DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(ORP)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Catalytic mechanism of the mismatch-specific DNA glycosylase methyl-CpG-binding domain 4.
Biochem.J., 477, 2020
|
|
