7F36
 
 | | TacT complexed with acetyl-glycyl-tRNAGly | | Descriptor: | ACETYLAMINO-ACETIC ACID, CALCIUM ION, N-acetyltransferase domain-containing protein, ... | | Authors: | Yashiro, Y, Tomita, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Molecular basis of glycyl-tRNAGly acetylation by TacT from Salmonella Typhimurium
Cell Rep, 37, 2021
|
|
7F37
 
 | | Crystal structure of AtaT2-AtaR2 complex | | Descriptor: | DUF1778 domain-containing protein, GNAT family N-acetyltransferase | | Authors: | Yashiro, Y, Tomita, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Molecular basis of glycyl-tRNAGly acetylation by TacT from Salmonella Typhimurium
Cell Rep, 37, 2021
|
|
8J6V
 
 | |
6AJN
 
 | | Crystal structure of AtaTR bound with AcCoA | | Descriptor: | ACETYL COENZYME *A, DUF1778 domain-containing protein, N-acetyltransferase | | Authors: | Yashiro, Y, Yamashita, S, Tomita, K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal Structure of the Enterohemorrhagic Escherichia coli AtaT-AtaR Toxin-Antitoxin Complex.
Structure, 27, 2019
|
|
6AJM
 
 | | Crystal structure of apo AtaTR | | Descriptor: | DUF1778 domain-containing protein, N-acetyltransferase, TRIETHYLENE GLYCOL | | Authors: | Yashiro, Y, Yamashita, S, Tomita, K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structure of the Enterohemorrhagic Escherichia coli AtaT-AtaR Toxin-Antitoxin Complex.
Structure, 27, 2019
|
|
4U72
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (A260G mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
4U74
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (G262A mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
4YN8
 
 | | Crystal Structure of Response Regulator ChrA in Heme-Sensing Two Component System | | Descriptor: | MAGNESIUM ION, Response regulator ChrA, SULFATE ION | | Authors: | Doi, A, Nakamura, H, Shiro, Y, Sugimoto, H. | | Deposit date: | 2015-03-09 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the response regulator ChrA in the haem-sensing two-component system of Corynebacterium diphtheriae.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3CV9
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R73A/R84A mutant) in complex with 1alpha,25-dihydroxyvitamin D3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
3CV8
 
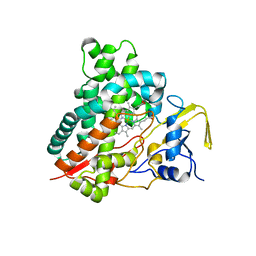 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84F mutant) | | Descriptor: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
5GUX
 
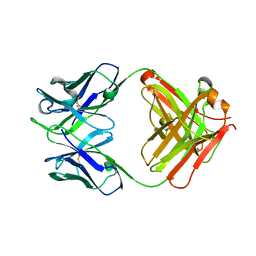 | | Cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with xenon | | Descriptor: | Antibody fab fragment heavy chain, Antibody fab fragment light chain, CALCIUM ION, ... | | Authors: | Ishii, S, Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GUW
 
 | | Complex of Cytochrome cd1 Nitrite Reductase and Nitric Oxide Reductase in Denitrification of Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6FWF
 
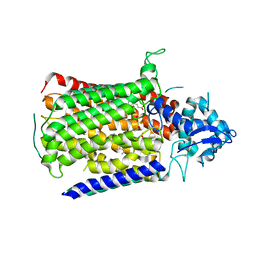 | | Low resolution structure of Neisseria meningitidis qNOR | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Young, D, Antonyuk, S, Tosha, T, Hisano, T, Hasnain, S, Shiro, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme.
Sci Rep, 8, 2018
|
|
6QQ5
 
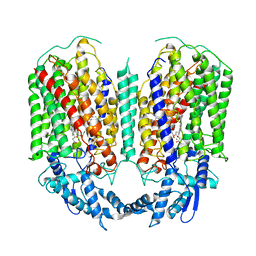 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric oxide reductase subunit B, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
6QQ6
 
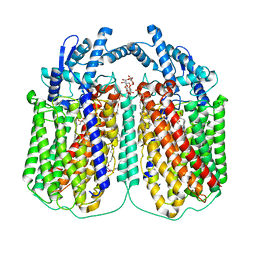 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) Val495Ala mutant from Alcaligenes xylosoxidans | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
3O0R
 
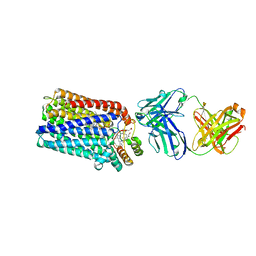 | | Crystal structure of nitric oxide reductase from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, FE (III) ION, HEME C, ... | | Authors: | Hino, T, Matsumoto, Y, Nagano, S, Sugimoto, H, Fukumori, Y, Murata, T, Iwata, S, Shiro, Y. | | Deposit date: | 2010-07-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of biological N2O generation by bacterial nitric oxide reductase
Science, 330, 2010
|
|
1EW0
 
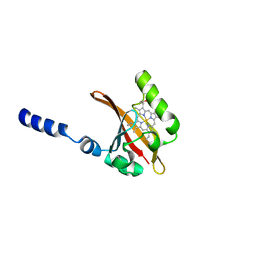 | | CRYSTAL STRUCTURE ANALYSIS OF THE SENSOR DOMAIN OF RMFIXL(FERROUS FORM) | | Descriptor: | FIXL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 2000-04-21 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
8ZGP
 
 | |
1JFB
 
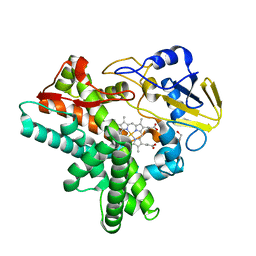 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferric resting state at atomic resolution | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, nitric-oxide reductase cytochrome P450 55A1 | | Authors: | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JFC
 
 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferrous CO state at atomic resolution | | Descriptor: | CARBON MONOXIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1CMJ
 
 | |
1CL6
 
 | |
5XHL
 
 | | Crystal Structure of HasAp with Gallium Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, Phthalocyanine containing GA | | Authors: | Shoji, O, Shisaka, Y, Iwai, Y, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HasAp with Gallium Phthalocyanine
to be published
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5L
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the absence of NADH (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
