7D24
 
 | | Hsp90 alpha N-terminal domain in complex with a 4B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D26
 
 | | Hsp90 alpha N-terminal domain in complex with a 8 compund | | Descriptor: | 6-chloranyl-9-[(2-phenyl-1,3-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D22
 
 | | Hsp90 alpha N-terminal domain in complex with a 6B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D1V
 
 | | Hsp90 alpha N-terminal domain in complex with a 6C compund | | Descriptor: | 6-chloranyl-9-[(3-propan-2-yl-1,2-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D25
 
 | | Hsp90 alpha N-terminal domain in complex with a 14 compund | | Descriptor: | 6-chloranyl-9-[(4-methylphenyl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
8ITN
 
 | | Crystal structure of USP47apo catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 47, ZINC ION | | Authors: | Kim, E.E, Shin, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of USP47 reveals a hot spot for inhibitor design.
Commun Biol, 6, 2023
|
|
8ITP
 
 | |
5ZQM
 
 | |
5ZQL
 
 | |
7FGN
 
 | | The crystal structure of the FAF1 UBL1 | | Descriptor: | FAS-associated factor 1 | | Authors: | Kim, E.E, ParK, J.K, Shin, S.C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | The complex of Fas-associated factor 1 with Hsp70 stabilizes the adherens junction integrity by suppressing RhoA activation
J Mol Cell Biol, 14, 2022
|
|
7FGM
 
 | | The complex crystals structure of the FAF1 UBL1_L-Hsp70 NBD with ADP and phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAS-associated factor 1, Heat shock 70 kDa protein 1A, ... | | Authors: | Kim, E.E, ParK, J.K, Shin, S.C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The complex of Fas-associated factor 1 with Hsp70 stabilizes the adherens junction integrity by suppressing RhoA activation
J Mol Cell Biol, 14, 2022
|
|
5GVJ
 
 | | Structure of FabK (M276A) mutant from Thermotoga maritima | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [FMN], SODIUM ION | | Authors: | Kim, E.E, Shin, S.C, Ha, B.H, Moon, J.H. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of FabK from Thermotoga maritima.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5GVH
 
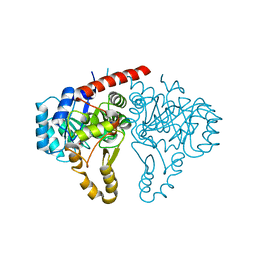 | | Structure of FabK from Thermotoga maritima | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [FMN], FLAVIN MONONUCLEOTIDE, SODIUM ION | | Authors: | Kim, E.E, Shin, S.C, Ha, B.H, Moon, J.H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural and biochemical characterization of FabK from Thermotoga maritima.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H22
 
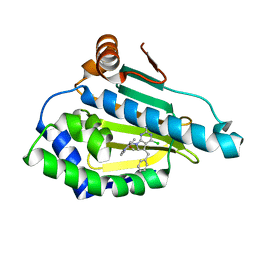 | | Hsp90 alpha N-terminal domain in complex with an inhibitor | | Descriptor: | 4-chloranyl-7-[(4-methoxy-3,5-dimethyl-pyridin-2-yl)methyl]-5-(phenylmethyl)pyrrolo[2,3-d]pyrimidin-2-amine, Hsp90aa1 protein | | Authors: | Kim, E.E, Shin, S.C, Keum, G.C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Synthesis and in vitro antiproliferative activity of C5-benzyl substituted 2-amino-pyrrolo[2,3-d]pyrimidines as potent Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5X3S
 
 | |
7TBA
 
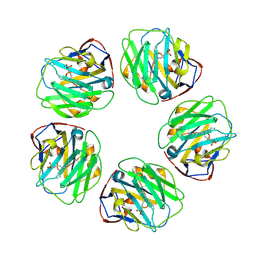 | | Pentraxin - ligand complex | | Descriptor: | C-reactive protein, CALCIUM ION, [3-(dibutylamino)propyl]phosphonic acid | | Authors: | Shing, K.S.C.T, Morton, C.J, Parker, M.W. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A novel phosphocholine-mimetic inhibits a pro-inflammatory conformational change in C-reactive protein.
Embo Mol Med, 15, 2023
|
|
6LVP
 
 | |
7XRH
 
 | | Feruloyl esterase from Lactobacillus acidophilus | | Descriptor: | Cinnamoyl esterase | | Authors: | Hwang, J, Lee, C.W, Lee, J.H, Do, H. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7XRI
 
 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7YC0
 
 | | Acetylesterase (LgEstI) W.T. | | Descriptor: | ACETATE ION, Alpha/beta hydrolase, CHLORIDE ION | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
7YC4
 
 | | Acetylesterase (LgEstI) F207A | | Descriptor: | Alpha/beta hydrolase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
6LVO
 
 | |
7DVN
 
 | | Crystal structure of a MarR family protein in complex with a lipid-like effector molecule from the psychrophilic bacterium Paenisporosarcina sp. TG-14 | | Descriptor: | MarR family transcriptional regulator, PALMITIC ACID | | Authors: | Lee, C.W, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a MarR family protein from the psychrophilic bacterium Paenisporosarcina sp. TG-14 in complex with a lipid-like molecule.
Iucrj, 8, 2021
|
|
5JCL
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Os09g0567300 protein | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCK
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Os09g0567300 protein | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6
|
|
