4S0X
 
 | | Structure of three phase partition - treated lipase from Thermomyces lanuginosa in complex with lauric acid at 2.1 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, ... | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of three phase partition - treated lipase from Thermomyces lanuginosa in complex with lauric acid at 2.1 A resolution
To be Published
|
|
4S0M
 
 | | Crystal Structure of nucleoside diphosphate kinase at 1.92 A resolution from acinetobacter baumannii | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Sikarwar, J, Shukla, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-01-02 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Crystal Structure of nucleoside diphosphate kinase at 1.92 A resolution from Acinetobacter baumannii
To be Published
|
|
4Y55
 
 | | Crystal structure of Buffalo lactoperoxidase with Rhodanide at 2.09 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Gupta, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Buffalo lactoperoxidase with Rhodanide at 2.09 Angstrom resolution
To Be Published
|
|
4ZC1
 
 | |
4ZGB
 
 | | Structure of untreated lipase from Thermomyces lanuginosa at 2.3 A resolution | | Descriptor: | Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-04-22 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhancement of stability of a lipase by subjecting to three phase partitioning (TPP): structures of native and TPP-treated lipase from Thermomyces lanuginosa
Sustain Chem Process, 2015
|
|
4DXV
 
 | | Crystal structure of Dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Mg and Cl ions at 1.80 A resolution | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Kumar, M, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Mg and Cl ions at 1.80 A resolution
To be Published
|
|
5B6P
 
 | | Structure of the dodecameric type-II dehydrogenate dehydratase from Acinetobacter baumannii at 2.00 A resolution | | Descriptor: | 3-dehydroquinate dehydratase, SULFATE ION | | Authors: | Kumar, M, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding studies and structure determination of the recombinantly produced type-II 3-dehydroquinate dehydratase from Acinetobacter baumannii.
Int. J. Biol. Macromol., 94, 2017
|
|
5Y98
 
 | | Crystal structure of native unbound peptidyl tRNA hydrolase from Acinetobacter baumannii at 1.36 A resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Iqbal, N, Singh, N, Kaushik, S, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-23 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Search of multiple hot spots on the surface of peptidyl-tRNA hydrolase: structural, binding and antibacterial studies.
Biochem. J., 475, 2018
|
|
5Y9A
 
 | | Crystal structure of the complex of peptidyl tRNA hydrolase with a phosphate ion at the substrate binding site and cytarabine at a new ligand binding site at 1.1 A resolution | | Descriptor: | CYTARABINE, PHOSPHATE ION, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Iqbal, N, Singh, N, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-23 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Search of multiple hot spots on the surface of peptidyl-tRNA hydrolase: structural, binding and antibacterial studies.
Biochem. J., 475, 2018
|
|
1U4J
 
 | | Crystal structure of a carbohydrate induced dimer of group I phospholipase A2 from Bungarus caeruleus at 2.1 A resolution | | Descriptor: | ACETIC ACID, CHLORIDE ION, SODIUM ION, ... | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of a carbohydrate induced homodimer of phospholipase A(2) from Bungarus caeruleus at 2.1A resolution
J.Struct.Biol., 149, 2005
|
|
1OXR
 
 | | Aspirin induces its Anti-inflammatory effects through its specific binding to Phospholipase A2: Crystal structure of the complex formed between Phospholipase A2 and Aspirin at 1.9A resolution | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Singh, R.K, Ethayathulla, A.S, Jabeen, T, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2003-04-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Aspirin induces its anti-inflammatory effects through its specific binding to phospholipase A2: crystal structure of the complex formed between phospholipase A2 and aspirin at 1.9 angstroms resolution.
J.Drug Target., 13, 2005
|
|
1OYF
 
 | | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol | | Descriptor: | 6-METHYLHEPTAN-1-OL, ACETIC ACID, Phospholipase A2, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-04-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol
To be Published
|
|
1OWS
 
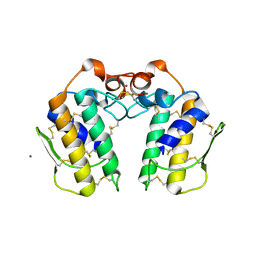 | | Crystal structure of a C49 Phospholipase A2 from Indian cobra reveals carbohydrate binding in the hydrophobic channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Phospholipase A2, ZINC ION | | Authors: | Jabeen, T, Jasti, J, Singh, N, Singh, R.K, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a C49 Phospholipase A2 from Indian cobra reveals carbohydrate binding in the hydrophobic channel
To be Published
|
|
1OWQ
 
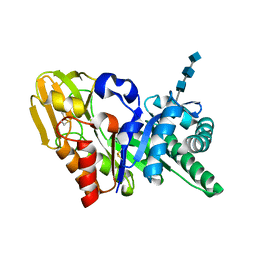 | | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Kumar, J, Sharma, S, Jasti, J, Bhushan, A, Singh, T.P. | | Deposit date: | 2003-03-29 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution
To be Published
|
|
5UNH
 
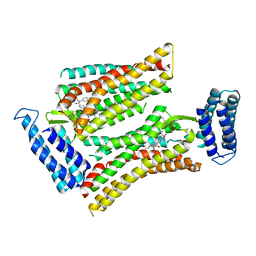 | | Synchrotron structure of human angiotensin II type 2 receptor in complex with compound 2 (N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'- biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide) | | Descriptor: | N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide, Soluble cytochrome b562,Type-2 angiotensin II receptor | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
3NNO
 
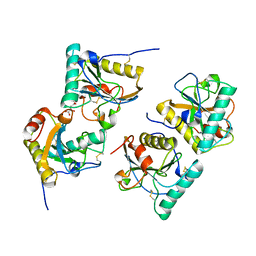 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1, alpha-L-rhamnopyranose | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution
To be Published
|
|
5UNG
 
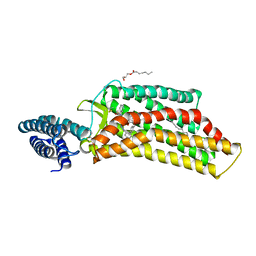 | | XFEL structure of human angiotensin II type 2 receptor (Orthorhombic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl] methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide, ... | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
5UNF
 
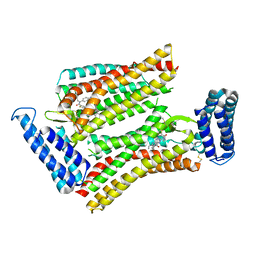 | | XFEL structure of human angiotensin II type 2 receptor (Monoclinic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]) | | Descriptor: | Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
1PO8
 
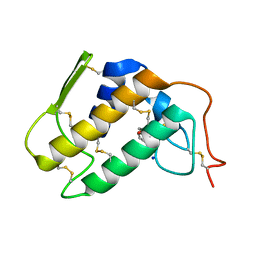 | | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution. | | Descriptor: | HEPTANOIC ACID, Phospholipase A2, SODIUM ION | | Authors: | Singh, G, Jayasankar, J, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-06-14 | | Release date: | 2004-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution.
To be Published
|
|
1Q6V
 
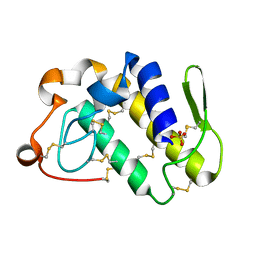 | | First crystal structure of a C49 monomer PLA2 from the venom of Daboia russelli pulchella at 1.8 A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Pal, A, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-14 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | First crystal structure of a C49 PLA2 from the venom of Daboia russelli pulchella at 1.8A resolution
To be Published
|
|
5WRF
 
 | | Crystal structure of dodecameric type II dehydroquinate dehydratase from Acinetobacter baumannii with unexplained connecting electron density between free cysteine residues of molecular pairs | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase | | Authors: | Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-01 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of dodecameric type II dehydroquinate dehydratase from Acinetobacter baumannii with unexplained connecting electron density between free cysteine residues of molecular pairs
To Be Published
|
|
5WUY
 
 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution | | Descriptor: | Chorismate synthase | | Authors: | Iqbal, N, Chaudhary, A, Shukla, K.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution
To Be Published
|
|
5WV3
 
 | | Crystal structure of bovine lactoperoxidase with a partial Glu258-heme linkage at 2.07 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of activation of mammalian heme peroxidases
Prog. Biophys. Mol. Biol., 133, 2018
|
|
5X47
 
 | |
3QS0
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-19 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel AT 2.5 A resolution
To be Published
|
|
