7MDY
 
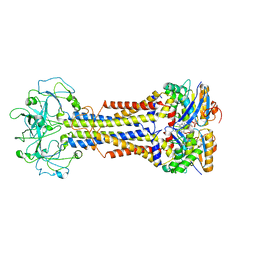 | | LolCDE nucleotide-bound | | Descriptor: | ADP ORTHOVANADATE, Lipo-releasing system transmembrane protein lolC, Lipoprotein transporter subunit LolE, ... | | Authors: | Sharma, S, Liao, M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins
Nat Commun, 12, 2021
|
|
7MDX
 
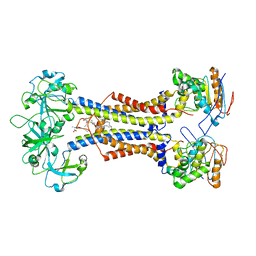 | | LolCDE nucleotide-free | | Descriptor: | (2R)-2-(tridecanoyloxy)propyl hexadecanoate, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, ... | | Authors: | Sharma, S, Liao, M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins
Nat Commun, 12, 2021
|
|
8FKJ
 
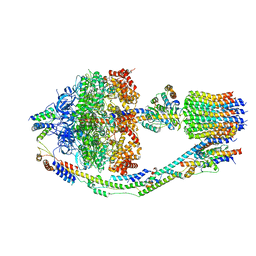 | | Yeast ATP Synthase in conformation-3, at pH 6 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FL8
 
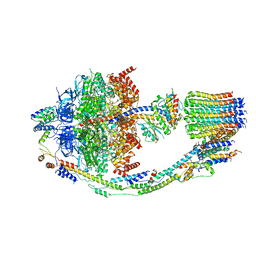 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F39
 
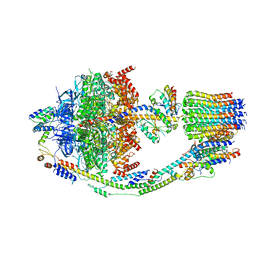 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F29
 
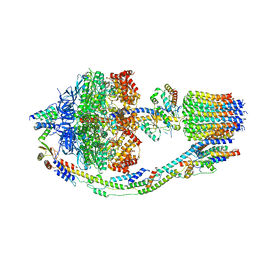 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7SHX
 
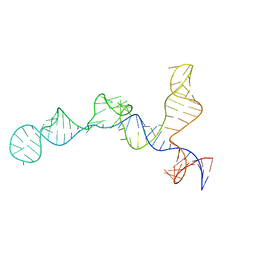 | |
8U1E
 
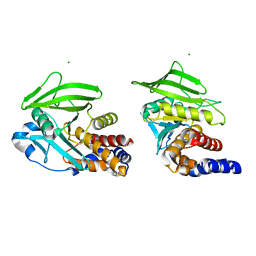 | |
6W3M
 
 | |
1NKX
 
 | | CRYSTAL STRUCTURE OF A PROTEOLYTICALLY GENERATED FUNCTIONAL MONOFERRIC C-LOBE OF BOVINE LACTOFERRIN AT 1.9A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Sharma, S, Jasti, J, Kumar, J, Mohanty, A.K, Singh, T.P. | | Deposit date: | 2003-01-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Proteolytically Generated Functional Monoferric C-lobe of
Bovine Lactoferrin at 1.9A Resolution
J.Mol.Biol., 331, 2003
|
|
2YIM
 
 | | The enolisation chemistry of a thioester-dependent racemase: the 1.4 A crystal structure of a complex with a planar reaction intermediate analogue | | Descriptor: | 2-METHYLACETOACETYL COA, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sharma, S, Bhaumik, P, Venkatesan, R, Hiltunen, J.K, Conzelmann, E, Juffer, A.H, Wierenga, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The Enolization Chemistry of a Thioester-Dependent Racemase: The 1.4 A Crystal Structure of a Reaction Intermediate Complex Characterized by Detailed Qm/Mm Calculations.
J Phys Chem B, 116, 2012
|
|
6U79
 
 | |
2MDL
 
 | |
8DU7
 
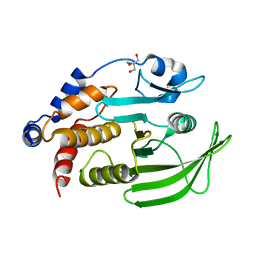 | | Room-temperature serial synchrotron crystallography (SSX) structure of apo PTP1B | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Sharma, S, Keedy, D.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room-temperature serial synchrotron crystallography of the human phosphatase PTP1B.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8IL9
 
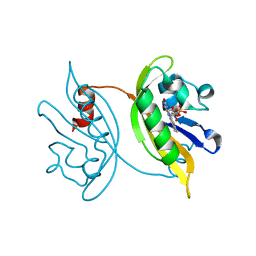 | | Crystal structure of the LOV1 Q122N mutant of Klebsormidium nitens phototropin | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gautam, A.K, Sharma, S, Gourinath, S, Kateriya, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Unusual photodynamic characteristics of the light-oxygen-voltage domain of phototropin linked to terrestrial adaptation of Klebsormidium nitens.
Febs J., 291, 2024
|
|
8J68
 
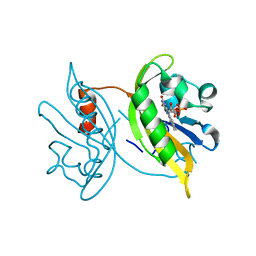 | | Crystal structure of the LOV1 R60K mutant of Klebsormidium nitens phototropin | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gautam, A.K, Sharma, S, Gourinath, S, Kateriya, S. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Unusual photodynamic characteristics of the light-oxygen-voltage domain of phototropin linked to terrestrial adaptation of Klebsormidium nitens.
Febs J., 291, 2024
|
|
8IYN
 
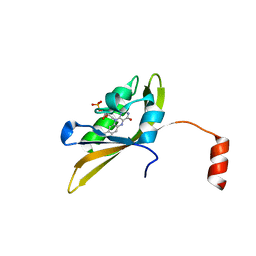 | | Crystal structure of LOV1 D33N mutant of phototropin from Klebsormidium nitens | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gautam, A.K, Sharma, S, Gourinath, S, Kateriya, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Unusual photodynamic characteristics of the light-oxygen-voltage domain of phototropin linked to terrestrial adaptation of Klebsormidium nitens.
Febs J., 291, 2024
|
|
8I11
 
 | | Crystal structure of LOV1 domain of phototropin from Klebsormidium nitens | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gautam, A.K, Sharma, S, Gourinath, S, Kateriya, S. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Unusual photodynamic characteristics of the light-oxygen-voltage domain of phototropin linked to terrestrial adaptation of Klebsormidium nitens.
Febs J., 291, 2024
|
|
6IF9
 
 | |
2JVD
 
 | | Solution NMR structure of the folded N-terminal fragment of UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384-1-46 | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Sharma, S, Huang, Y.J, Zhao, L, Owens, L.A, Stokes, K, Jiang, M, Xiao, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis.
Proteins, 72, 2008
|
|
1Q7A
 
 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
1PO8
 
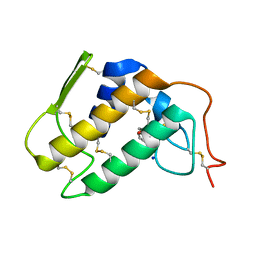 | | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution. | | Descriptor: | HEPTANOIC ACID, Phospholipase A2, SODIUM ION | | Authors: | Singh, G, Jayasankar, J, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-06-14 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution.
To be Published
|
|
3T39
 
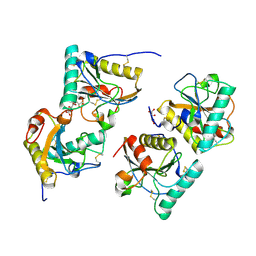 | | Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution
To be Published
|
|
3GCI
 
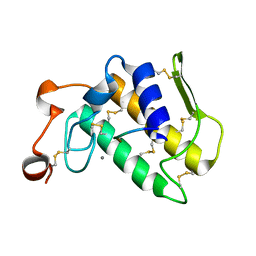 | | Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution | | Descriptor: | CALCIUM ION, Heptapeptide from Amyloid beta A4 protein, Phospholipase A2 isoform 3 | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution
To be Published
|
|
9UGE
 
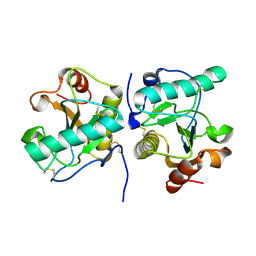 | | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution | | Descriptor: | D-MALATE, OXALIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Barik, D, Ahmad, N, Maurya, A, Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution
To Be Published
|
|
