5KUF
 
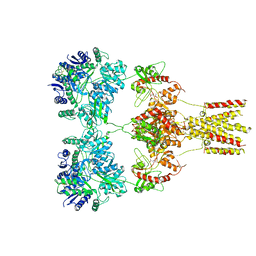 | | GluK2EM with 2S,4R-4-methylglutamate | | Descriptor: | 2S,4R-4-METHYLGLUTAMATE, Glutamate receptor ionotropic, kainate 2 | | Authors: | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5KUH
 
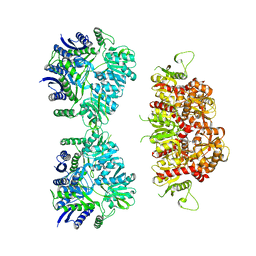 | | GluK2EM with LY466195 | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | Authors: | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5ICT
 
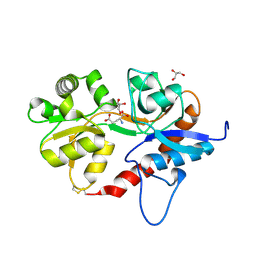 | |
5LLQ
 
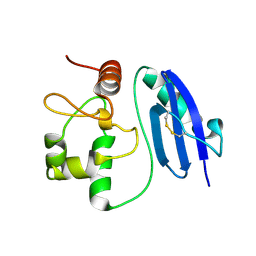 | |
4WXD
 
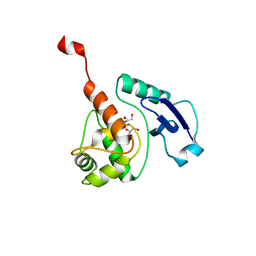 | | Crystal structure of Mycobacterium tuberculosis OGT-R37K | | Descriptor: | GLYCEROL, Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Miggiano, R, Rossi, F, Garavaglia, S, Rizzi, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis O6-methylguanine-DNA methyltransferase protein clusters assembled on to damaged DNA.
Biochem.J., 473, 2016
|
|
4WX9
 
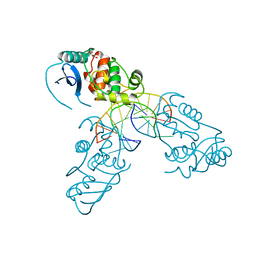 | | Crystal structure of Mycobacterium tuberculosis OGT in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*TP*GP*(E1X)P*CP*TP*AP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3'), GLYCEROL, ... | | Authors: | Miggiano, R, Rossi, F, Garavaglia, S, Rizzi, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis O6-methylguanine-DNA methyltransferase protein clusters assembled on to damaged DNA.
Biochem.J., 473, 2016
|
|
4WXC
 
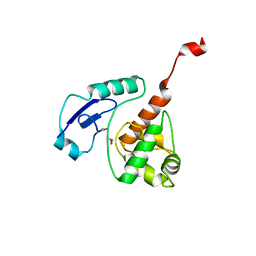 | | Crystal structure of Mycobacterium tuberculosis OGT-Y139F | | Descriptor: | 1,2-ETHANEDIOL, Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Miggiano, R, Rossi, F, Garavaglia, S, Rizzi, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis O6-methylguanine-DNA methyltransferase protein clusters assembled on to damaged DNA.
Biochem.J., 473, 2016
|
|
4ZYE
 
 | |
4ZYG
 
 | |
4ZYH
 
 | |
4ZYD
 
 | |
4WXJ
 
 | | Drosophila muscle GluRIIB complex with glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor IIB,Glutamate receptor IIB | | Authors: | Dharkar, P, Mayer, M.L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Functional reconstitution of Drosophila melanogaster NMJ glutamate receptors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DT6
 
 | |
5DTB
 
 | |
5EHM
 
 | |
5EHS
 
 | |
5CMM
 
 | |
5CMK
 
 | | Crystal structure of the GluK2EM LBD dimer assembly complex with glutamate and LY466195 | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Chittori, S, Mayer, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
