4LUK
 
 | |
4LTA
 
 | |
2CFX
 
 | | Structure of B.subtilis LrpC | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR LRPC | | Authors: | Thaw, P, Rafferty, J.B. | | Deposit date: | 2006-02-24 | | Release date: | 2006-03-28 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into Gene Transcriptional Regulation and Effector Binding by the Lrp/Asnc Family.
Nucleic Acids Res., 34, 2006
|
|
6EZM
 
 | | Imidazoleglycerol-phosphate dehydratase from Saccharomyces cerevisiae | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2R)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EZJ
 
 | | Imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FWH
 
 | | Acanthamoeba IGPD in complex with R-C348 to 1.7A resolution | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Roberts, C.W, Bisson, C, Baker, P.J. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional studies of histidine biosynthesis in Acanthamoeba spp. demonstrates a novel molecular arrangement and target for antimicrobials.
PLoS ONE, 13, 2018
|
|
6H2D
 
 | |
6GRK
 
 | |
6GRJ
 
 | | Structure of the AhlB pore of the tripartite alpha-pore forming toxin, AHL, from Aeromonas hydrophila. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AhlB, CHLORIDE ION, ... | | Authors: | Churchill-Angus, A.M, Wilson, J.S, Baker, P.J. | | Deposit date: | 2018-06-11 | | Release date: | 2019-07-03 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Identification and structural analysis of the tripartite alpha-pore forming toxin of Aeromonas hydrophila.
Nat Commun, 10, 2019
|
|
6H2E
 
 | |
6H2F
 
 | | Structure of the pre-pore AhlB of the tripartite alpha-pore forming toxin, AHL, from Aeromonas hydrophila. | | Descriptor: | AhlB, PHOSPHATE ION | | Authors: | Churchill-Angus, A.M, Wilson, J.S, Baker, P.J. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification and structural analysis of the tripartite alpha-pore forming toxin of Aeromonas hydrophila.
Nat Commun, 10, 2019
|
|
1GTM
 
 | | STRUCTURE OF GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Yip, K.S.P, Stillman, T.J, Britton, K.L, Pasquo, A, Rice, D.W. | | Deposit date: | 1996-08-22 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
1AUP
 
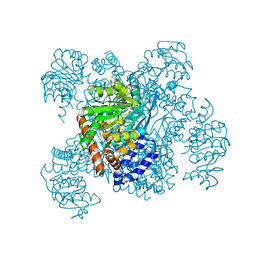 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Baker, P.J, Waugh, M.L, Stillman, T.J, Turnbull, A.P, Rice, D.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determinants of substrate specificity in the superfamily of amino acid dehydrogenases.
Biochemistry, 36, 1997
|
|
1BGV
 
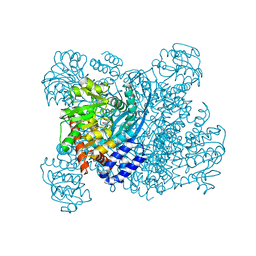 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, GLUTAMIC ACID | | Authors: | Stillman, T.J, Baker, P.J, Britton, K.L, Rice, D.W. | | Deposit date: | 1998-06-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis.
J.Mol.Biol., 234, 1993
|
|
1F8G
 
 | | THE X-RAY STRUCTURE OF NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE FROM RHODOSPIRILLUM RUBRUM COMPLEXED WITH NAD+ | | Descriptor: | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Buckley, P.A, Baz Jackson, J, Schneider, T, White, S.A, Rice, D.W, Baker, P.J. | | Deposit date: | 2000-06-30 | | Release date: | 2001-06-30 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-protein recognition, hydride transfer and proton pumping in the transhydrogenase complex.
Structure Fold.Des., 8, 2000
|
|
1HRD
 
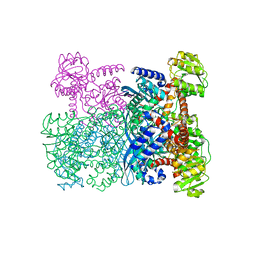 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Britton, K.L, Baker, P.J, Stillman, T.J, Rice, D.W. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
1QJH
 
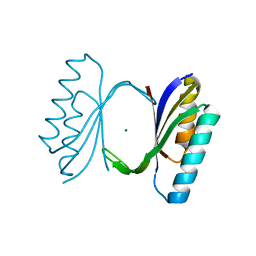 | |
2B5W
 
 | | Crystal structure of D38C glucose dehydrogenase mutant from Haloferax mediterranei | | Descriptor: | CITRATE ANION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B5V
 
 | | Crystal structure of glucose dehydrogenase from Haloferax mediterranei | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, glucose dehydrogenase | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2BXJ
 
 | | Double Mutant of the Ribosomal Protein S6 | | Descriptor: | 30S RIBOSOMAL PROTEIN S6 | | Authors: | Otzen, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antagonism, Non-Native Interactions and Non-Two-State Folding in S6 Revealed by Double-Mutant Cycle Analysis.
Protein Eng.Des.Sel., 18, 2005
|
|
1CQN
 
 | |
1CQM
 
 | |
1EDO
 
 | | THE X-RAY STRUCTURE OF BETA-KETO ACYL CARRIER PROTEIN REDUCTASE FROM BRASSICA NAPUS COMPLEXED WITH NADP+ | | Descriptor: | BETA-KETO ACYL CARRIER PROTEIN REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fisher, M, Kroon, J.T, Martindale, W, Stuitje, A.R, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2000-01-28 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray structure of Brassica napus beta-keto acyl carrier protein reductase and its implications for substrate binding and catalysis.
Structure Fold.Des., 8, 2000
|
|
1FKA
 
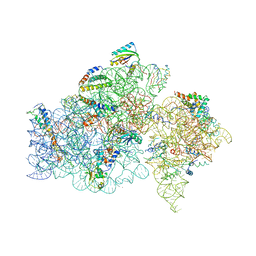 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
1H7Y
 
 | | Translationally Controlled Tumor-associated Protein p23fyp from Schizosaccharomyces pombe | | Descriptor: | TRANSLATIONALLY CONTROLLED TUMOR PROTEIN | | Authors: | Thaw, P, Baxter, N.J, Sedelnikova, S.E, Price, C, Waltho, J.P, Craven, C.J. | | Deposit date: | 2001-01-19 | | Release date: | 2001-08-09 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones.
Nat. Struct. Biol., 8, 2001
|
|
