6S4A
 
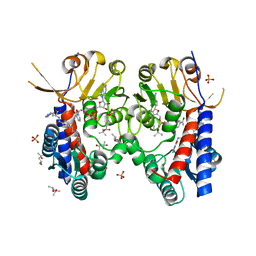 | | Structure of human MTHFD2 in complex with TH9028 | | Descriptor: | (2~{S})-2-[[5-[[2,4-bis(azanyl)-6-oxidanylidene-5~{H}-pyrimidin-5-yl]carbamoylamino]pyridin-2-yl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4F
 
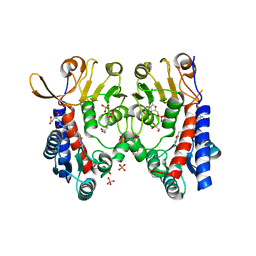 | | Structure of human MTHFD2 in complex with TH9619 | | Descriptor: | (E,4S)-4-[[5-[2-[2,6-bis(azanyl)-4-oxidanylidene-1H-pyrimidin-5-yl]ethanoylamino]-3-fluoranyl-pyridin-2-yl]carbonylamino]pent-2-enedioic acid, ADENOSINE-5'-DIPHOSPHATE, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Scaletti, E.R, Gustafsson, R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4E
 
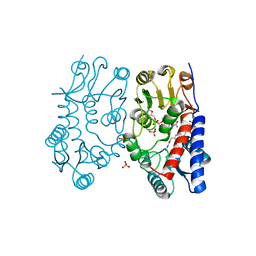 | | Structure of human MTHFD2 in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
5NGS
 
 | | Crystal structure of human MTH1 in complex with inhibitor 6-[(2-phenylethyl)sulfanyl]-7H-purin-2-amine | | Descriptor: | 6-(2-phenylethylsulfanyl)-7~{H}-purin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
5NGR
 
 | | Crystal structure of human MTH1 in complex with fragment inhibitor 8-(methylsulfanyl)-7H-purin-6-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-methylsulfanyl-7~{H}-purin-6-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
5NGT
 
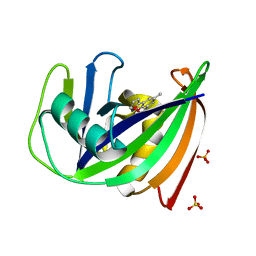 | | Crystal structure of human MTH1 in complex with inhibitor 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
8CEY
 
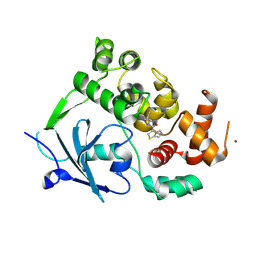 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233. | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-(3,3-dimethylcyclobutyl)imidazo[5,1-b][1,3]thiazole-3-carboxamide | | Authors: | Kosenina, S, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
8CEX
 
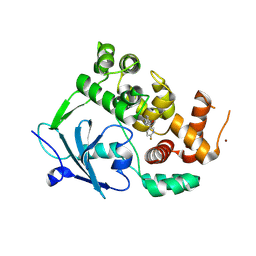 | |
7PZ1
 
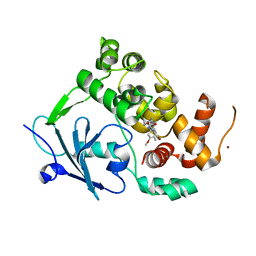 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH8535 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-bromanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(3-methoxy-4-methyl-phenyl)piperidine-1-carboxamide, GLYCEROL, ... | | Authors: | Scaletti, E.R, Helleday, T, Stenmark, P. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
7QEL
 
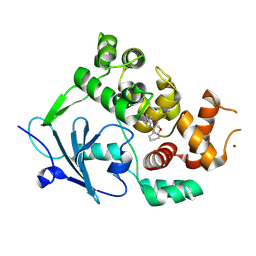 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011247 | | Descriptor: | (2~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)spiro[2.3]hexane-2-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
4N1U
 
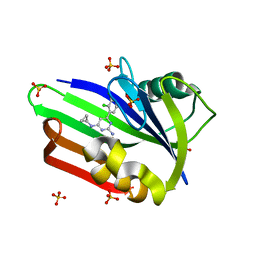 | | Structure of human MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
4N1T
 
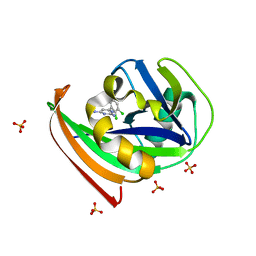 | | Structure of human MTH1 in complex with TH287 | | Descriptor: | 6-(2,3-dichlorophenyl)-N~4~-methylpyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
6G40
 
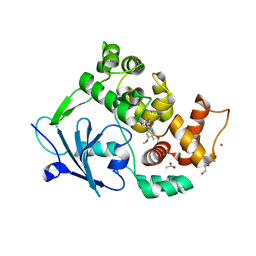 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH9525 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
6G3X
 
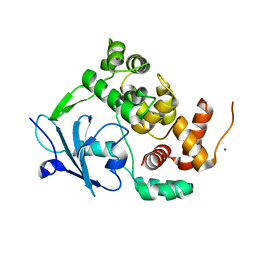 | |
6G3Y
 
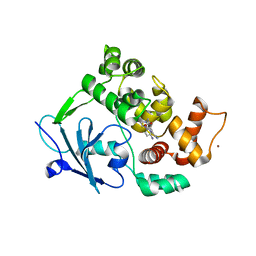 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH5675 | | Descriptor: | 4-(4-azanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(4-iodophenyl)piperidine-1-carboxamide, ACETATE ION, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.
Science, 362, 2018
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
6RLW
 
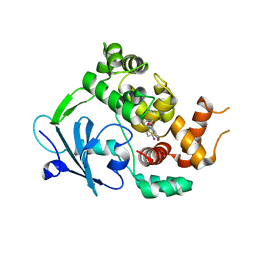 | |
5NQR
 
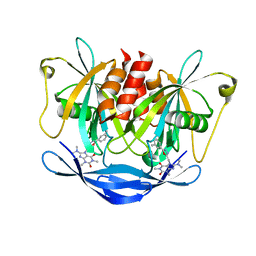 | | Potent inhibitors of NUDT5 silence hormone signaling in breast cancer | | Descriptor: | 8-(dimethylamino)-1,3-dimethyl-7-[[5-(3-methylphenyl)-1,3,4-oxadiazol-2-yl]methyl]purine-2,6-dione, ADP-sugar pyrophosphatase | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2017-04-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells.
Nat Commun, 9, 2018
|
|
7ZC7
 
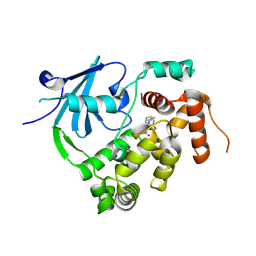 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-25 | | Release date: | 2023-07-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
7ZG3
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228 | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-[(1~{S})-1,2,2-trimethylcyclopropyl]pyrrolo[1,2-c]pyrimidine-3-carboxamide | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-04-01 | | Release date: | 2023-08-16 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
5NWH
 
 | | Potent inhibitors of NUDT5 silence hormone signaling in breast cancer | | Descriptor: | 7-[[5-(3,4-dichlorophenyl)-1,3,4-oxadiazol-2-yl]methyl]-1,3-dimethyl-8-piperazin-1-yl-purine-2,6-dione, ADP-sugar pyrophosphatase | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2017-05-06 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells.
Nat Commun, 9, 2018
|
|
7Z3Y
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)-2,6-bis(fluoranyl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Scaletti, E.R, Stenmark, P. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
7Z5B
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013546 | | Descriptor: | 2-[4-(2,4-dimethyl-1~{H}-imidazol-5-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
7Z5R
 
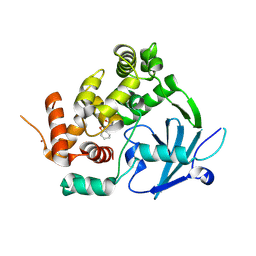 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012035 | | Descriptor: | (7~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)bicyclo[4.2.0]octa-1,3,5-triene-7-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-22 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
7AYZ
 
 | |
