4JG9
 
 | | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis | | Descriptor: | Lipoprotein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis
To be Published
|
|
4DD5
 
 | | Biosynthetic Thiolase (ThlA1) from Clostridium difficile | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biosynthetic Thiolase (ThlA1) from Clostridium difficile
To be Published
|
|
4JID
 
 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | Descriptor: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
4EBK
 
 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, tobramycin-bound | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, tobramycin-bound
To be Published
|
|
4JJP
 
 | | 2.06 Angstrom resolution crystal structure of phosphomethylpyrimidine kinase (thiD)from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Phosphomethylpyrimidine kinase | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | 2.06 Angstrom resolution crystal structure of phosphomethylpyrimidine kinase (thiD)from Clostridium difficile 630
To be Published
|
|
4EW5
 
 | | C-terminal domain of inner membrane protein CigR from Salmonella enterica. | | Descriptor: | 1,2-ETHANEDIOL, CigR Protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Niemann, G.S, Merkley, E.D, Savchenko, A, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | C-terminal domain of inner membrane protein CigR from Salmonella enterica.
To be Published
|
|
2KKX
 
 | | Solution Structure of C-terminal domain of reduced NleG2-3 (residues 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
2KKY
 
 | | Solution Structure of C-terminal domain of oxidized NleG2-3 (residue 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
4EUY
 
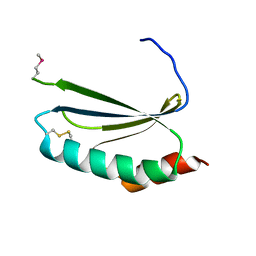 | | Crystal structure of thioredoxin-like protein BCE_0499 from Bacillus cereus ATCC 10987 | | Descriptor: | Uncharacterized protein | | Authors: | Shabalin, I.G, Kagan, O, Chruszcz, M, Grabowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of thioredoxin-like protein BCE_0499 from
Bacillus cereus
To be Published
|
|
4KI3
 
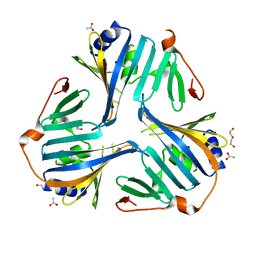 | | 1.70 Angstrom resolution crystal structure of outer-membrane lipoprotein carrier protein (lolA) from Yersinia pestis CO92 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-05-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom resolution crystal structure of outer-membrane lipoprotein carrier protein (lolA) from Yersinia pestis CO92
To be Published
|
|
4LAT
 
 | | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Phosphate-binding protein PstS 1 | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate
To be Published
|
|
4G65
 
 | | Potassium transporter peripheral membrane component (trkA) from Vibrio vulnificus | | Descriptor: | CHLORIDE ION, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-18 | | Release date: | 2012-08-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Potassium transporter peripheral membrane component (trkA) from Vibrio vulnificus.
To be Published
|
|
4GKH
 
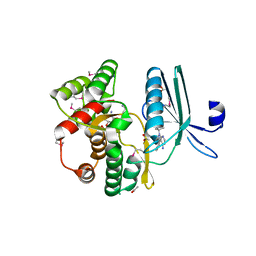 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4MFG
 
 | | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile.
TO BE PUBLISHED
|
|
4GKI
 
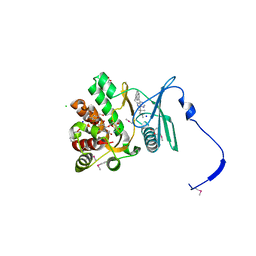 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NM-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4MHB
 
 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
2AV9
 
 | | Crystal Structure of the PA5185 protein from Pseudomonas Aeruginosa Strain PAO1. | | Descriptor: | SULFATE ION, Thioesterase | | Authors: | Chruszcz, M, Wang, S, Cymborowski, M, Kudritska, M, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-29 | | Release date: | 2005-10-18 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
4MPH
 
 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-bound | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0301 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
4GYT
 
 | | Crystal structure of lpg0076 protein from Legionella pneumophila | | Descriptor: | SODIUM ION, uncharacterized protein | | Authors: | Michalska, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Crystal structure of lpg0076 protein from Legionella pneumophila
To be Published
|
|
4MUT
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUS
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Ala-D-Ala phosphinate analog | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MYO
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
2P35
 
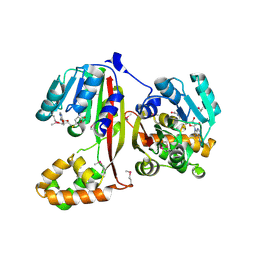 | | Crystal structure of trans-aconitate methyltransferase from Agrobacterium tumefaciens | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Trans-aconitate 2-methyltransferase | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of trans-aconitate methyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2OV9
 
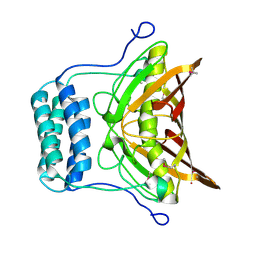 | | Crystal structure of protein RHA08564, thioesterase superfamily protein | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-13 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein RHA08564, thioesterase superfamily protein
To be Published
|
|
2PQQ
 
 | | Structural Genomics, the crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | FORMIC ACID, Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2)
To be Published
|
|
