4WBS
 
 | |
8FBI
 
 | |
8FBJ
 
 | |
8FBN
 
 | |
8FBO
 
 | |
8FBK
 
 | |
4WV9
 
 | | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane | | Descriptor: | (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J.M, Sankaran, B. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound.
To Be Published
|
|
8G9J
 
 | |
8G9K
 
 | |
8GA6
 
 | |
8GA7
 
 | |
8GEL
 
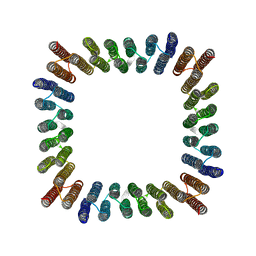 | | Cryo-EM structure of synthetic tetrameric building block sC4 | | Descriptor: | sC4 | | Authors: | Redler, R.L, Huddy, T.F, Hsia, Y, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Blueprinting extendable nanomaterials with standardized protein blocks.
Nature, 627, 2024
|
|
7RKC
 
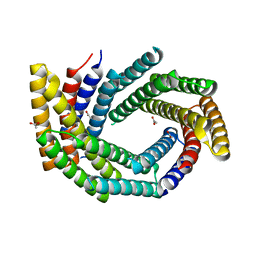 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633 | | Descriptor: | ACETATE ION, D_3_633 | | Authors: | Kennedy, M.A, Stoddard, B.L, Hicks, D.R, Bera, A.K. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4OPA
 
 | |
8CYK
 
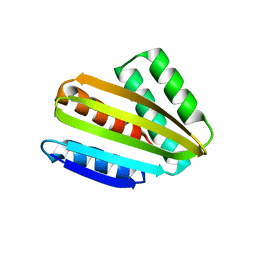 | | Crystal structure of hallucinated protein HALC1_878 | | Descriptor: | HALC1_878 | | Authors: | Ragotte, R.J, Bera, A.K, Milles, L.F, Wicky, B.I.M, Baker, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Robust deep learning-based protein sequence design using ProteinMPNN.
Science, 378, 2022
|
|
3LRF
 
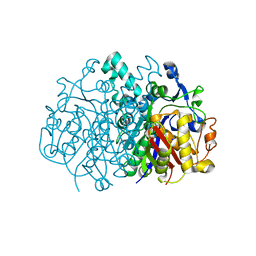 | |
5U0C
 
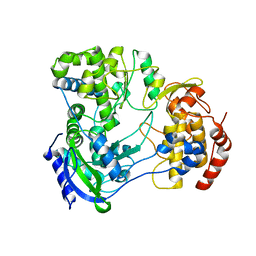 | |
5U0B
 
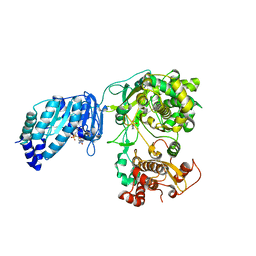 | | Structure of full-length Zika virus NS5 | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Zhao, B, Du, F. | | Deposit date: | 2016-11-23 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the Zika virus full-length NS5 protein.
Nat Commun, 8, 2017
|
|
5JEK
 
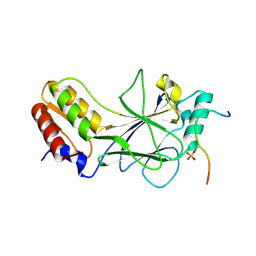 | | Phosphorylated MAVS in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, MAVS peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEM
 
 | | Complex of IRF-3 with CBP | | Descriptor: | CREB-binding protein, Interferon regulatory factor 3 | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JER
 
 | | Structure of Rotavirus NSP1 bound to IRF-3 | | Descriptor: | Interferon regulatory factor 3, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEL
 
 | | Phosphorylated TRIF in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, Phosphorylated TRIF peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEO
 
 | | Phosphorylated Rotavirus NSP1 in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, PHOSPHATE ION, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6CYA
 
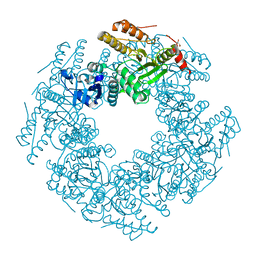 | | Rotavirus SA11 NSP2 S313A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 2 | | Authors: | Anish, R, Hu, L, Prasad, B.V.V. | | Deposit date: | 2018-04-05 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation cascade regulates the formation and maturation of rotaviral replication factories.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4Y0V
 
 | |
