4XX3
 
 | |
7RRO
 
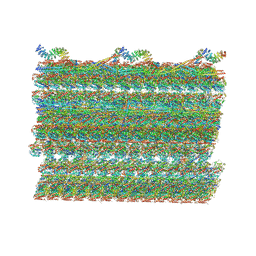 | | Structure of the 48-nm repeat doublet microtubule from bovine tracheal cilia | | Descriptor: | Armadillo repeat containing 4, Chromosome 3 C1orf194 homolog, Cilia and flagella associated protein 161, ... | | Authors: | Gui, M, Anderson, J.R, Botsch, J.J, Meleppattu, S, Singh, S.K, Zhang, Q, Brown, A. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo identification of mammalian ciliary motility proteins using cryo-EM.
Cell, 184, 2021
|
|
8GJA
 
 | | RAD51C-XRCC3 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8GJ9
 
 | |
8GJ8
 
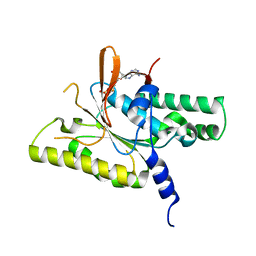 | | RAD51C C-terminal domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAD51C | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8GOG
 
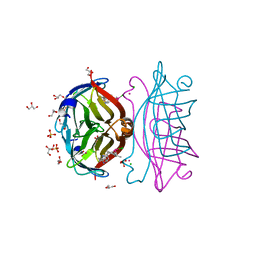 | | Structure of streptavidin mutant (S112Y-K121E) complexed with biotin-cyclopentadienyl-rhodium (III)(Cp*-Rh(III)) | | Descriptor: | CHLORIDE ION, GLYCEROL, RHODIUM(III) ION, ... | | Authors: | Sairaman, A, Mukherjee, P, Maiti, D, Bhaumik, P. | | Deposit date: | 2022-08-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Artificial metalloenzyme catalyzed enantiodivergent synthesis of isoindolones
NAT SYNTH
|
|
4OR1
 
 | | Structure and mechanism of fibronectin binding and biofilm formation of enteroaggregative Escherischia coli AAF fimbriae | | Descriptor: | ACETATE ION, Invasin homolog AafB, Major fimbrial subunit of aggregative adherence fimbria II AafA chimeric construct, ... | | Authors: | Lee, W.-C, Garnett, J.A, Yang, Y, Matthews, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
7TDM
 
 | |
7TDN
 
 | |
7PWB
 
 | | dTDP-sugar epimerase from Coxiella burnetii in complex with dTDP | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PVI
 
 | | dTDP-sugar epimerase | | Descriptor: | CITRATE ANION, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.434 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PWH
 
 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7PWI
 
 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, dTDP-4-keto-rhamnose 3,5-epimerase,dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.326 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
4PHX
 
 | |
4PH8
 
 | |
5KH0
 
 | |
5LAD
 
 | |
5JHQ
 
 | |
7RK8
 
 | |
7RK9
 
 | |
3OD8
 
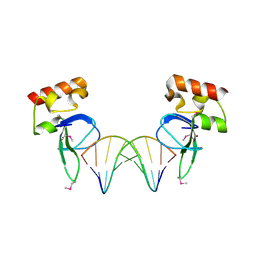 | | Human PARP-1 zinc finger 1 (Zn1) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*AP*GP*CP*GP*GP*C)-3', 5'-D(*GP*CP*CP*GP*CP*TP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
3ODA
 
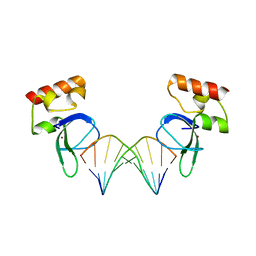 | | Human PARP-1 zinc finger 1 (Zn1) bound to DNA | | Descriptor: | 5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3', Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
3ODE
 
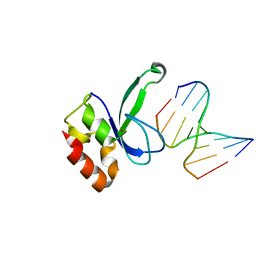 | | Human PARP-1 zinc finger 2 (Zn2) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*AP*GP*CP*G)-3', 5'-D(*CP*GP*CP*TP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
3ODC
 
 | | Human PARP-1 zinc finger 2 (Zn2) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*GP*AP*CP*G)-3', 5'-D(*CP*GP*TP*CP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
2ZHX
 
 | | Crystal structure of Uracil-DNA Glycosylase from Mycobacterium tuberculosis in complex with a proteinaceous inhibitor | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Kaushal, P.S, Talawar, R.K, Krishna, P.D.V, Varshney, U, Vijayan, M. | | Deposit date: | 2008-02-11 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unique features of the structure and interactions of mycobacterial uracil-DNA glycosylase: structure of a complex of the Mycobacterium tuberculosis enzyme in comparison with those from other sources
Acta Crystallogr.,Sect.D, 64, 2008
|
|
