9F5V
 
 | | Crystal structure of thiol peroxidase from Helicobacter pylori (HpTx, reduced) | | Descriptor: | Thiol peroxidase | | Authors: | Fiedler, M.K, Gong, R, Fuchs, S, Rox, K, Friedrich, V, Pfeiffer, D, Reinhardt, T, Mibus, C, Huber, M, Hess, C, Mejias-Luque, R, Gerhard, M, Groll, M, Sieber, S.A. | | Deposit date: | 2024-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 5-Nitroimidazole ethers boost anti-Helicobacter pylori activity via a dual mode of action and effectively eradicate infections in vivo
to be published
|
|
9F64
 
 | | Crystal structure of thiol peroxidase mutant (C94A) in complex with Metro* (H. pylori | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-azanyl-2-methyl-imidazol-1-yl)ethanol, SODIUM ION, ... | | Authors: | Fiedler, M.K, Gong, R, Fuchs, S, Rox, K, Friedrich, V, Pfeiffer, D, Reinhardt, T, Mibus, C, Huber, M, Hess, C, Mejias-Luque, R, Gerhard, M, Groll, M, Sieber, S.A. | | Deposit date: | 2024-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 5-Nitroimidazole ethers boost anti-Helicobacter pylori activity via a dual mode of action and effectively eradicate infections in vivo
to be published
|
|
9F65
 
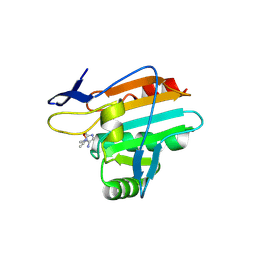 | | Crystal structure of thiol peroxidase mutant (C94A) in complex with Metro-P3* (H. pylori) | | Descriptor: | 3-methyl-2-(prop-2-ynoxymethyl)imidazol-4-amine, Thiol peroxidase | | Authors: | Fiedler, M.K, Gong, R, Fuchs, S, Rox, K, Friedrich, V, Pfeiffer, D, Reinhardt, T, Mibus, C, Huber, M, Hess, C, Mejias-Luque, R, Gerhard, M, Groll, M, Sieber, S.A. | | Deposit date: | 2024-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 5-Nitroimidazole ethers boost anti-Helicobacter pylori activity via a dual mode of action and effectively eradicate infections in vivo
to be published
|
|
8CRB
 
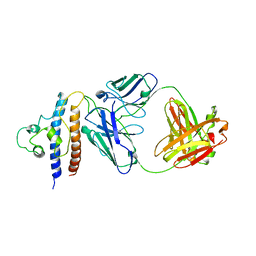 | | Cryo-EM structure of PcrV/Fab(11-E5) | | Descriptor: | Heavy chain, Light chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
8CR9
 
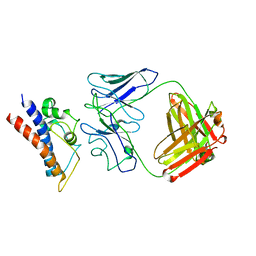 | | Cryo-EM structure of PcrV/Fab(30-B8) | | Descriptor: | Heavy chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV, light chain | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
7P4U
 
 | |
7ZOC
 
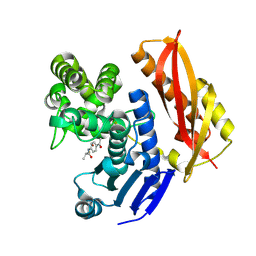 | |
8A4T
 
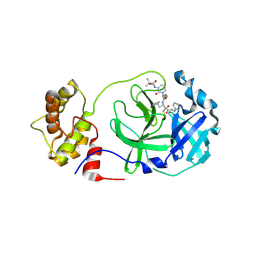 | | crystal structures of diastereomer (S,S,S)-13b (13b-K) in complex with the SARS-CoV-2 Mpro | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
8A4Q
 
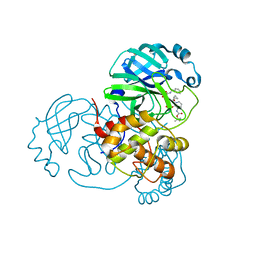 | | crystal structures of diastereomer (R,S,S)-13b (13b-H) in complex with the SARS-CoV-2 Mpro. | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
9GV2
 
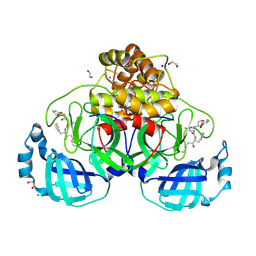 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor FP237 (compound 8p in publication) | | Descriptor: | (2S)-2-[2-(3-methoxyphenoxy)ethanoylamino]-4-methyl-N-[(2S)-3-oxidanylidene-1-phenyl-pentan-2-yl]pentanamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Strater, N, Sylvester, K, Muller, C.E, Flury, P, Kruger, N, Breidenbach, J, Guetschow, M, Laufer, S.A, Pillaiyar, T. | | Deposit date: | 2024-09-20 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, Synthesis, and Unprecedented Interactions of Covalent Dipeptide-Based Inhibitors of SARS-CoV-2 Main Protease and Its Variants Displaying Potent Antiviral Activity.
J.Med.Chem., 68, 2025
|
|
9F2X
 
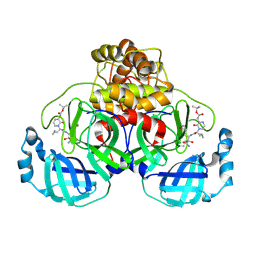 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR03 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[4-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-3-oxidanylidene-pyrazin-2-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9F2V
 
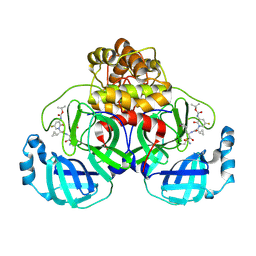 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR02 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{R},3~{S})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-5-fluoranyl-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9F3A
 
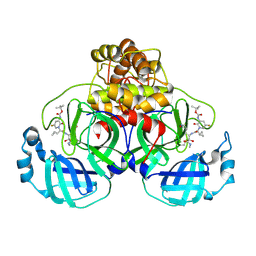 | | Crystal structure of SARS-CoV-2 Mpro in complex with RK-325 | | Descriptor: | 3C-like proteinase nsp5, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-5-fluoranyl-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9F39
 
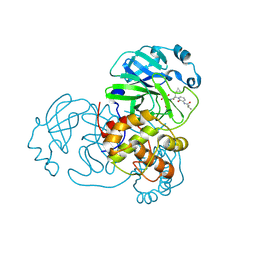 | | Crystal structure of SARS-CoV-2 Mpro in complex with RK-54 | | Descriptor: | (2R,3R)-3-[[(2S)-3-cyclopropyl-2-[3-(2-methylpropanoylamino)-2-oxidanylidene-pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5 | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9GMQ
 
 | | Crystal structure of the Mpro of SARS COV-2 in complex with the MG-87 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-08-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9FHQ
 
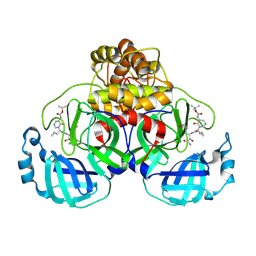 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR04 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-6-oxidanylidene-pyrimidin-5-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-05-28 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
8AIJ
 
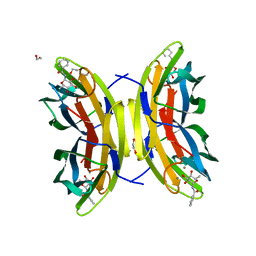 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PAO1 IN COMPLEX WITH N-(alpha-L-Fucopyranosyl)benzamide (6) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Meiers, J, Mala, P, Varrot, A, Siebs, E, Imberty, A, Titz, A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of N -beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB.
J.Med.Chem., 65, 2022
|
|
8AIY
 
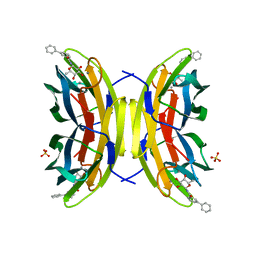 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PAO1 IN COMPLEX WITH N-(beta-L-Fucopyranosyl)-biphenyl-3-carboxamide (4i) | | Descriptor: | CALCIUM ION, Fucose-binding lectin PA-IIL, N-(beta-L-Fucopyranosyl)-biphenyl-3-carboxamide, ... | | Authors: | Meiers, J, Mala, P, Varrot, A, Siebs, E, Imberty, A, Titz, A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of N -beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB.
J.Med.Chem., 65, 2022
|
|
8PHI
 
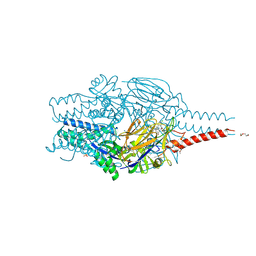 | | Crystal structure of prefusion-stabilized RSV F Variant DS-Cav1 in complex with Lonafarnib | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Rajak, M, Krey, T. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor.
Nat Commun, 15, 2024
|
|
8R1B
 
 | |
5MAY
 
 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PA14 IN COMPLEX WITH 2-Thiophenesulfonamide-N-(beta-L-fucopyranosyl methyl) | | Descriptor: | CALCIUM ION, Fucose-binding lectin PA-IIL, N-methyl-2-thiophenesulfonamide, ... | | Authors: | Sommer, R, Imberty, A, Titz, A, Varrot, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Glycomimetic, Orally Bioavailable LecB Inhibitors Block Biofilm Formation of Pseudomonas aeruginosa.
J. Am. Chem. Soc., 140, 2018
|
|
8AIV
 
 | | Mpro of SARS COV-2 in complex with the MG-100 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
8AIU
 
 | | Mpro of SARS COV-2 in complex with the MG-97 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
8AJ0
 
 | | Mpro of SARS COV-2 in complex with the RK-90 inhibitor | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(3-phenylpropanoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
8AIZ
 
 | | Mpro of SARS-CoV-2 in complex with the RK-68 inhibitor | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-3-cyclopropyl-2-[2-oxidanylidene-3-(2-phenylethanoylamino)pyridin-1-yl]propanoyl]amino]-~{N}-methyl-2-oxidanyl-4-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butanamide, CHLORIDE ION, Replicase polyprotein 1ab | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
