4B0N
 
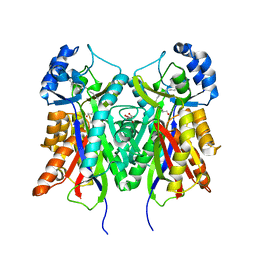 | | Crystal structure of PKS-I from the brown algae Ectocarpus siliculosus | | Descriptor: | ARACHIDONIC ACID, MALONIC ACID, POLYKETIDE SYNTHASE III | | Authors: | Leroux, C, Meslet-Cladiere, L, Delage, L, Goulitquer, S, Leblanc, C, Ar Gall, E, Stiger-Pouvreau, V, Potin, P, Czjzek, M. | | Deposit date: | 2012-07-03 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure/Function Analysis of a Type III Polyketide Synthase in the Brown Alga Ectocarpus Siliculosus Reveals a Biochemical Pathway in Phlorotannin Monomer Biosynthesis.
Plant Cell, 25, 2013
|
|
5K3V
 
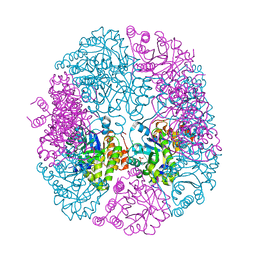 | | apo-PDX1.3 (Arabidopsis) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | Deposit date: | 2016-05-20 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2Z
 
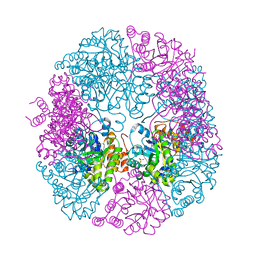 | | PDX1.3-adduct (Arabidopsis) | | Descriptor: | 1,2-ETHANEDIOL, 2-azanylpenta-1,4-dien-3-one, CHLORIDE ION, ... | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6HXG
 
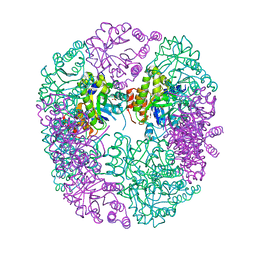 | | PDX1.2/PDX1.3 complex (intermediate) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6HX3
 
 | | PDX1.2/PDX1.3 complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6HYE
 
 | | PDX1.2/PDX1.3 complex (PDX1.3:K97A) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4H67
 
 | | Crystal structure of HMP synthase Thi5 from S. cerevisiae | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyrimidine precursor biosynthesis enzyme THI5, SULFATE ION | | Authors: | Coquille, S.C, Roux, C, Fitzpatrick, T, Thore, S. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Last Piece in the Vitamin B1 Biosynthesis Puzzle: STRUCTURAL AND FUNCTIONAL INSIGHT INTO YEAST 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE PHOSPHATE (HMP-P) SYNTHASE.
J.Biol.Chem., 287, 2012
|
|
4H65
 
 | |
4H6D
 
 | | Crystal structure of PLP-soaked HMP synthase Thi5 from S. cerevisiae | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyrimidine precursor biosynthesis enzyme THI5 | | Authors: | Coquille, S.C, Roux, C, Fitzpatrick, T, Thore, S. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Last Piece in the Vitamin B1 Biosynthesis Puzzle: STRUCTURAL AND FUNCTIONAL INSIGHT INTO YEAST 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE PHOSPHATE (HMP-P) SYNTHASE.
J.Biol.Chem., 287, 2012
|
|
4N7Q
 
 | | Crystal structure of eukaryotic THIC from A. thaliana | | Descriptor: | COBALT (II) ION, HEXANE-1,6-DIOL, Phosphomethylpyrimidine synthase, ... | | Authors: | Coquille, S.C, Roux, C, Mehta, A, Begley, T.P, Fitzpatrick, T.B, Thore, S. | | Deposit date: | 2013-10-16 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution crystal structure of the eukaryotic HMP-P synthase (THIC) from Arabidopsis thaliana.
J.Struct.Biol., 184, 2013
|
|
2XK8
 
 | | Structure of Nek2 bound to aminopyrazine compound 15 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-2-methoxybenzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XK6
 
 | | Structure of Nek2 bound to aminopyrazine compound 36 | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2, cis-4-[3-amino-6-(3-cyclopropylthiophen-2-yl)pyrazin-2-yl]cyclohexanecarboxylic acid | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XNP
 
 | | Structure of Nek2 bound to CCT244858 | | Descriptor: | 1,2-ETHANEDIOL, 4-{5-[(1-METHYLPIPERIDIN-4-YL)OXY]-1H-BENZIMIDAZOL-1-YL}-2-{(1R)-1-[2-(TRIFLUOROMETHYL)PHENYL]ETHOXY}BENZAMIDE, CHLORIDE ION, ... | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Benzimidazole Inhibitors Induce a Dfg-Out Conformation of Never in Mitosis Gene A-Related Kinase 2 (Nek2) without Binding to the Back Pocket and Reveal a Nonlinear Structure-Activity Relationship.
J.Med.Chem., 54, 2011
|
|
2XK7
 
 | | Structure of Nek2 bound to aminopyrazine compound 23 | | Descriptor: | (3R,4R)-1-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-3-ethylpiperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XKD
 
 | | Structure of Nek2 bound to aminopyrazine compound 12 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]benzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XKE
 
 | | Structure of Nek2 bound to Aminipyrazine Compound 5 | | Descriptor: | 1-[3-amino-6-(3-methoxyphenyl)pyrazin-2-yl]piperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XKF
 
 | | Structure of Nek2 bound to aminopyrazine compound 2 | | Descriptor: | 1-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]piperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
6SK9
 
 | | Nek2 bound to purine compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, GLYCEROL, Serine/threonine-protein kinase Nek2 | | Authors: | Bayliss, R, Byrne, M.J, Mas-Droux, C. | | Deposit date: | 2019-08-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
2C2G
 
 | |
2C2B
 
 | | Crystallographic structure of Arabidopsis thaliana Threonine synthase complexed with pyridoxal phosphate and S-adenosylmethionine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRIDOXAL-5'-PHOSPHATE, S-ADENOSYLMETHIONINE, ... | | Authors: | Mas-Droux, C, Biou, V, Dumas, R. | | Deposit date: | 2005-09-27 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric Threonine Synthase: Reorganization of the Pyridoxal Phosphate Site Upon Asymmetric Activation Through S-Adenosylmethionine Binding to a Novel Site.
J.Biol.Chem., 281, 2006
|
|
2CDQ
 
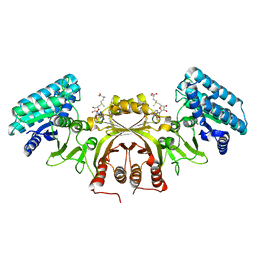 | | Crystal structure of Arabidopsis thaliana aspartate kinase complexed with lysine and S- adenosylmethionine | | Descriptor: | ASPARTOKINASE, D(-)-TARTARIC ACID, LYSINE, ... | | Authors: | Mas-Droux, C, Curien, G, Robert-Genthon, M, Laurencin, M, Ferrer, J.L, Dumas, R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Novel Organization of Act Domains in Allosteric Enzymes Revealed by the Crystal Structure of Arabidopsis Aspartate Kinase
Plant Cell, 18, 2006
|
|
2XK3
 
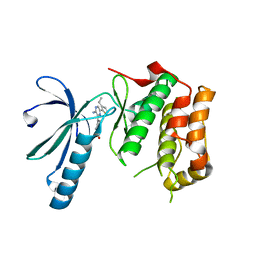 | | Structure of Nek2 bound to Aminopyrazine compound 35 | | Descriptor: | 4-[3-AMINO-6-(3-ETHYLTHIOPHEN-2-YL)PYRAZIN-2-YL]CYCLOHEXANE-1-CARBOXYLIC ACID, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XK4
 
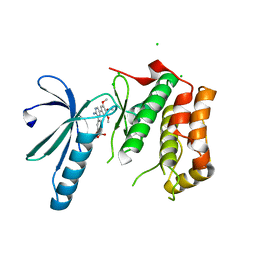 | | Structure of Nek2 bound to aminopyrazine compound 17 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-2-ethoxybenzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XNO
 
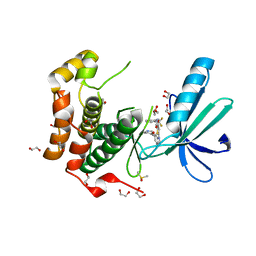 | | Structure of Nek2 bound to CCT243779 | | Descriptor: | 1,2-ETHANEDIOL, 5-{6-[(1-methylpiperidin-4-yl)oxy]-1H-benzimidazol-1-yl}-3-{[2-(trifluoromethyl)benzyl]oxy}thiophene-2-carboxamide, CHLORIDE ION, ... | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Benzimidazole Inhibitors Induce a Dfg-Out Conformation of Never in Mitosis Gene A-Related Kinase 2 (Nek2) without Binding to the Back Pocket and Reveal a Nonlinear Structure-Activity Relationship.
J.Med.Chem., 54, 2011
|
|
2XNM
 
 | | Structure of NEK2 bound to CCT | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-{6-[(1-METHYLPIPERIDIN-4-YL)OXY]-1H-BENZIMIDAZOL-1-YL}-3-{(1R)-1-[2-(TRIFLUOROMETHYL)PHENYL]ETHOXY}THIOPHENE-2-CARBOXAMIDE, ... | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Benzimidazole Inhibitors Induce a Dfg-Out Conformation of Never in Mitosis Gene A-Related Kinase 2 (Nek2) without Binding to the Back Pocket and Reveal a Nonlinear Structure-Activity Relationship.
J.Med.Chem., 54, 2011
|
|
