2KEJ
 
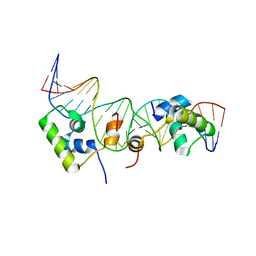 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEK
 
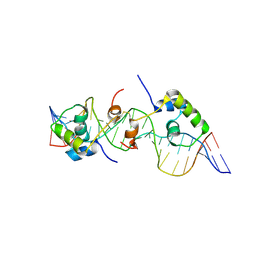 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O3 | | Descriptor: | DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*GP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*AP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*TP*GP*CP*GP*TP*TP*GP*CP*GP*CP*TP*CP*AP*CP*TP*GP*CP*CP*G)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEI
 
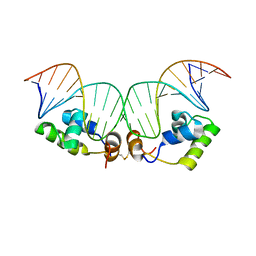 | | Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1 | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2BM2
 
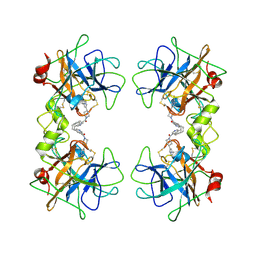 | | human beta-II tryptase in complex with 4-(3-Aminomethyl-phenyl)- piperidin-1-yl-(5-phenethyl- pyridin-3-yl)-methanone | | Descriptor: | 1-[3-(1-{[5-(2-PHENYLETHYL)PYRIDIN-3-YL]CARBONYL}PIPERIDIN-4-YL)PHENYL]METHANAMINE, HUMAN BETA2 TRYPTASE | | Authors: | Maignan, S, Guilloteau, J.-P, Dupuy, A, Levell, J, Astles, P, Eastwood, P, Cairns, J, Houille, O, Aldous, S, Merriman, G, Whiteley, B, Pribish, J, Czekaj, M, Liang, G, Davidson, J, Harrison, T, Morley, A, Watson, S, Fenton, G, Mccarthy, C, Romano, J, Mathew, R, Engers, D, Gardyan, M, Sides, K, Kwong, J, Tsay, J, Rebello, S, Shen, L, Wang, J, Luo, Y, Giardino, O, Lim, H.-K, Smith, K, Pauls, H. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of 4-(3-Aminomethylphenyl) Piperidinyl-1-Amides: Novel, Potent, Selective, and Orally Bioavailable Inhibitors of Bii Tryptase
Bioorg.Med.Chem., 13, 2005
|
|
8STW
 
 | | K384N HUMAN CYSTATHIONINE BETA-SYNTHASE (delta 411-551) | | Descriptor: | Cystathionine beta-synthase, K384N variant, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mascarenhas, R, Roman, J, Banerjee, R. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Disease-causing cystathionine beta-synthase linker mutations impair allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
4ZCD
 
 | | Renalase in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Silvaggi, N.R, Moran, G.R, Roman, J.V. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6605 Å) | | Cite: | Bacterial Renalase: Structure and Kinetics of an Enzyme with 2- and 6-Dihydro-beta-NAD(P) Oxidase Activity from Pseudomonas phaseolicola.
Biochemistry, 54, 2015
|
|
4ZCC
 
 | | Renalase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Silvaggi, N.R, Moran, G.R, Roman, J.V. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Bacterial Renalase: Structure and Kinetics of an Enzyme with 2- and 6-Dihydro-beta-NAD(P) Oxidase Activity from Pseudomonas phaseolicola.
Biochemistry, 54, 2015
|
|
6DXL
 
 | | Linked amidobenzimidazole STING agonist | | Descriptor: | 1,1'-(butane-1,4-diyl)bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1H-benzimidazole-5-carboxamide}, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6DXG
 
 | | amidobenzimidazole (ABZI) STING agonists | | Descriptor: | 2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1-[(2R)-2-hydroxy-2-phenylethyl]-1H-benzimidazole-5-carboxamide, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6UL8
 
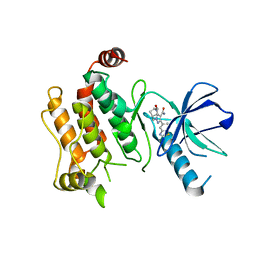 | | RIP2 kinase catalytic domain complex with (5S,6S,8R)-2-(benzo[d]thiazol-5-yl)-6-hydroxy-4,5,6,7,8,9-hexahydro-5,8-methanopyrazolo[1,5-a][1,3]diazocine-3-carboxamide | | Descriptor: | (5S,6S,8R)-2-(1,3-benzothiazol-5-yl)-6-hydroxy-4,5,6,7,8,9-hexahydro-5,8-methanopyrazolo[1,5-a][1,3]diazocine-3-carboxamide, CALCIUM ION, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Shewchuk, L.M, Convery, M.A. | | Deposit date: | 2019-10-07 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Pyrazolocarboxamides as Potent and Selective Receptor Interacting Protein 2 (RIP2) Kinase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6DCE
 
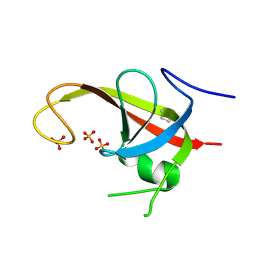 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
6SZE
 
 | |
6SZJ
 
 | |
6GMA
 
 | |
