3MX0
 
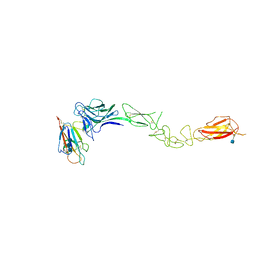 | | Crystal Structure of EphA2 ectodomain in complex with ephrin-A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 2, ... | | Authors: | Himanen, J.P, Yermekbayeva, L, Janes, P.W, Walker, J.R, Xu, K, Atapattu, L, Rajashankar, K.R, Mensinga, A, Lackmann, M, Nikolov, D.B, Dhe-Paganon, S. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1Q47
 
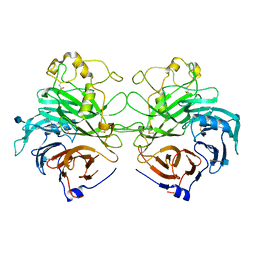 | | Structure of the Semaphorin 3A Receptor-Binding Module | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin 3A | | Authors: | Antipenko, A, Himanen, J.-P, van Leyen, K, Nardi-Dei, V, Lesniak, J, Barton, W.A, Rajashankar, K.R, Lu, M, Hoemme, C, Puschel, A, Nikolov, D. | | Deposit date: | 2003-08-01 | | Release date: | 2004-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the semaphorin-3A receptor binding module.
Neuron, 39, 2003
|
|
3OOC
 
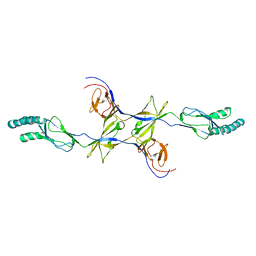 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2010-08-30 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli.
J.Mol.Biol., 393, 2009
|
|
1QZT
 
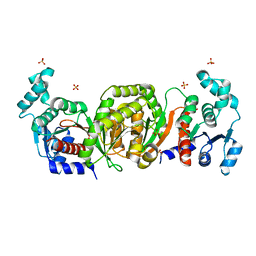 | | Phosphotransacetylase from Methanosarcina thermophila | | Descriptor: | Phosphate acetyltransferase, SULFATE ION | | Authors: | Iyer, P.P, Lawrence, S.H, Luther, K.B, Rajashankar, K.R, Yennawar, H.P, Ferry, J.G, Schindelin, H. | | Deposit date: | 2003-09-17 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of phosphotransacetylase from the methanogenic archaeon Methanosarcina thermophila.
STRUCTURE, 12, 2004
|
|
3QE5
 
 | | Complete structure of Streptococcus mutans Antigen I/II carboxy-terminus | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Major cell-surface adhesin PAc, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Crowley, P.J, Kelly, C, Mitchell, T.J, Brady, L.J, Deivanayagam, C. | | Deposit date: | 2011-01-19 | | Release date: | 2011-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the C-terminal Region of Streptococcus mutans Antigen I/II and Characterization of Salivary Agglutinin Adherence Domains.
J.Biol.Chem., 286, 2011
|
|
3R4F
 
 | | Prohead RNA | | Descriptor: | MAGNESIUM ION, pRNA | | Authors: | Ding, F, Lu, C, Zhano, W, Rajashankar, K.R, Anderson, D.L, Jardine, P.J, Grimes, S, Ke, A. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and assembly of the essential RNA ring component of a viral DNA packaging motor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4DS6
 
 | | Crystal structure of a group II intron in the pre-catalytic state | | Descriptor: | MAGNESIUM ION, Mutant Group IIC Intron | | Authors: | Chan, R.T, Robart, A.R, Rajashankar, K.R, Pyle, A.M, Toor, N. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.644 Å) | | Cite: | Crystal structure of a group II intron in the pre-catalytic state.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EN5
 
 | | Crystal structure of fluoride riboswitch, Tl-Acetate soaked | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4ENA
 
 | | Crystal structure of fluoride riboswitch, soaked in Cs+ | | Descriptor: | CESIUM ION, FLUORIDE ION, Fluoride riboswitch, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4ENB
 
 | | Crystal structure of fluoride riboswitch, bound to Iridium | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
1GQB
 
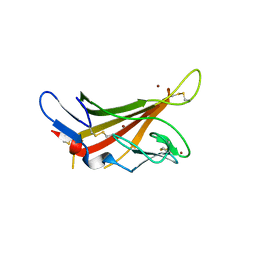 | | HUMAN MIR-RECEPTOR, REPEAT 11 | | Descriptor: | BROMIDE ION, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Dauter, M, Dauter, Z, Rajashankar, K.R, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2001-11-22 | | Release date: | 2002-12-05 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HA7
 
 | | STRUCTURE OF A LIGHT-HARVESTING PHYCOBILIPROTEIN, C-PHYCOCYANIN FROM SPIRULINA PLATENSIS AT 2.2A RESOLUTION | | Descriptor: | C-PHYCOCYANIN ALPHA CHAIN, C-PHYCOCYANIN BETA CHAIN, PHYCOCYANOBILIN | | Authors: | Padyana, A.K, Rajashankar, K.R, Ramakumar, S. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Light-Harvesting Protein C-Phycocyanin from Spirulina Platensis
Biochem.Biophys.Res.Commun., 282, 2001
|
|
1IIN
 
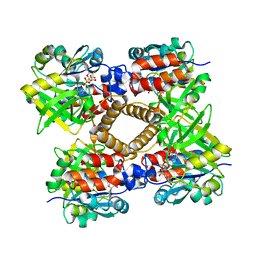 | | thymidylyltransferase complexed with UDP-glucose | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, glucose-1-phosphate thymidylyltransferase | | Authors: | Barton, W.A, Lesniak, J, Biggins, J.B, Jeffrey, P.D, Jiang, J, Rajashankar, K.R, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization.
Nat.Struct.Biol., 8, 2001
|
|
1IIM
 
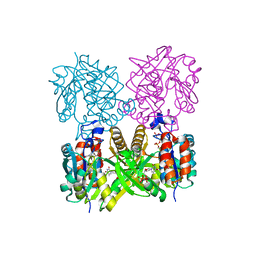 | | thymidylyltransferase complexed with TTP | | Descriptor: | THYMIDINE-5'-TRIPHOSPHATE, glucose-1-phosphate thymidylyltransferase | | Authors: | Barton, W.A, Lesniak, J, Biggins, J.B, Jeffrey, P.D, Jiang, J, Rajashankar, K.R, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization.
Nat.Struct.Biol., 8, 2001
|
|
1JLT
 
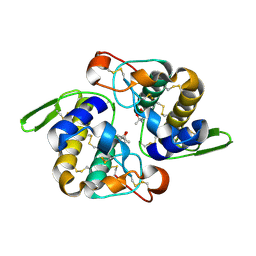 | | Vipoxin Complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOLIPASE A2, ... | | Authors: | Banumathi, S, Rajashankar, K.R, Notzel, C, Aleksiev, B, Singh, T.P, Genov, N, Betzel, C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the neurotoxic complex vipoxin at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1KGY
 
 | | Crystal Structure of the EphB2-ephrinB2 complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2, EPHRIN-B2 | | Authors: | Himanen, J.P, Rajashankar, K.R, Lackmann, M, Cowan, C.A, Henkemeyer, M, Nikolov, D.B. | | Deposit date: | 2001-11-28 | | Release date: | 2002-05-28 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an Eph receptor-ephrin complex.
Nature, 414, 2001
|
|
1KNZ
 
 | | Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer | | Descriptor: | 5'-R(*UP*GP*AP*CP*C)-3', Nonstructural RNA-binding Protein 34 | | Authors: | Deo, R.C, Groft, C.M, Rajashankar, K.R, Burley, S.K. | | Deposit date: | 2001-12-19 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer.
Cell(Cambridge,Mass.), 108, 2002
|
|
1LN3
 
 | | Structure of Human Phosphatidylcholine Transfer Protein in Complex with Palmitoyl-Linoleoyl Phosphatidylcholine (Seleno-Met Protein) | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
1LN1
 
 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with Dilinoleoylphosphatidylcholine | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
1LN2
 
 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with Dilinoleoylphosphatidylcholine (Seleno-Met Protein) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
4L3N
 
 | | Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Chen, Y, Rajashankar, K.R, Yang, Y, Agnihothram, S.S, Liu, C, Lin, Y.-L, Baric, R.S, Li, F. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the receptor-binding domain from newly emerged middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
4O6M
 
 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CMP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4O6N
 
 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CDP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4PTF
 
 | | Ternary crystal structure of yeast DNA polymerase epsilon with template G | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', ... | | Authors: | Jain, R, Rajashankar, K.R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Crystal Structure of Yeast DNA Polymerase epsilon Catalytic Domain.
Plos One, 9, 2014
|
|
1TLQ
 
 | | Crystal structure of protein ypjQ from Bacillus subtilis, Pfam DUF64 | | Descriptor: | CALCIUM ION, Hypothetical protein ypjQ | | Authors: | Kniewel, R, Rajashankar, K.R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Hypothetical Protein from Bacillus subtilis
To be Published
|
|
