3I6D
 
 | | Crystal structure of PPO from bacillus subtilis with AF | | Descriptor: | 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Shen, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis
J.Struct.Biol., 170, 2010
|
|
3NKS
 
 | | Structure of human protoporphyrinogen IX oxidase | | Descriptor: | 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Shen, Y. | | Deposit date: | 2010-06-21 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into human variegate porphyria disease
Faseb J., 25, 2011
|
|
6IGZ
 
 | | Structure of PSI-LHCI | | Descriptor: | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Xiong, P, Xiaochun, Q. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits.
Nat Plants, 5, 2019
|
|
8HOP
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Nap-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Nap-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HON
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Tyr-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Tyr-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOU
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Im-N-C4-Phe-Phe | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-N-C4-Phe-Phe, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOS
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4NO2)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4NO2)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOR
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4CH3)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4CH3)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOQ
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4CF3)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4CF3)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JC3
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-N-C4-Phe and hydroxylamine | | Descriptor: | 4-(pyridin-4-ylamino)butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
8JC4
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-Pid-Phe and hydroxylamine | | Descriptor: | 1-pyridin-4-ylpiperidine-4-carboxylic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
7Y0T
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanyl-L-phenylalanine | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, I7X-PHE-PHE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-06-06 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2R7I
 
 | | Crystal structure of catalytic subunit of protein kinase CK2 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
2R6M
 
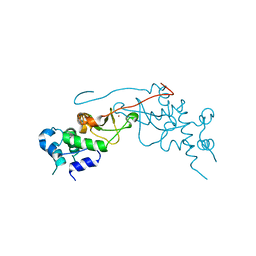 | | Crystal structure of rat CK2-beta subunit | | Descriptor: | Casein kinase II subunit beta, ZINC ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
1J4J
 
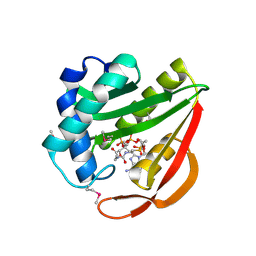 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
4IVO
 
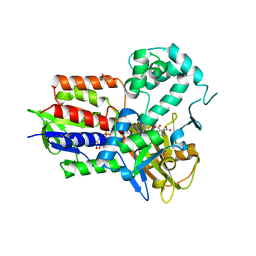 | | Structure of human protoporphyrinogen IX oxidase(R59Q) | | Descriptor: | 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiaohong, Q, Baifan, W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Quantitative structural insight into human variegate porphyria disease.
J.Biol.Chem., 288, 2013
|
|
4IVM
 
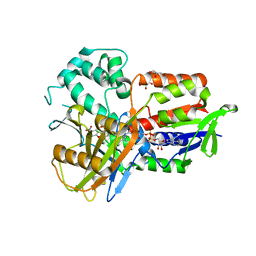 | | Structure of human protoporphyrinogen IX oxidase(R59G) | | Descriptor: | 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiaohong, Q, Baifan, W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Quantitative structural insight into human variegate porphyria disease.
J.Biol.Chem., 288, 2013
|
|
8SQF
 
 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SQG
 
 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
5GS4
 
 | | Crystal structure of estrogen receptor alpha in complex with a stabilized peptide antagonist | | Descriptor: | ARG-IAS-ILE-LEU-DNP-ARG-LEU-LEU-GLN, ESTRADIOL, Estrogen receptor, ... | | Authors: | Xie, M, Wang, T, Li, Z.-G. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-30 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Inhibition of ER alpha-Coactivator Interaction by High-Affinity N-Terminus Isoaspartic Acid Tethered Helical Peptides
J. Med. Chem., 60, 2017
|
|
5GTR
 
 | |
8HKG
 
 | |
7UR9
 
 | | SARS-Cov2 Main protease in complex with inhibitor CDD-1845 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[4-(methylamino)-4-oxobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
7US4
 
 | | Sars-Cov2 Main Protease in complex with CDD-1819 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M.M, Judge, A. | | Deposit date: | 2022-04-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
7URB
 
 | | Sars-Cov2 Main Protease in complex with CDD-1733 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-{(1S)-1-[4-(trifluoromethyl)phenyl]butyl}-1H-benzimidazole-7-carboxamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M.M, Judge, A. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
