6IQJ
 
 | |
6IQI
 
 | | crystal structure of Arabidopsis thaliana Profilin 2 | | Descriptor: | Profilin-2 | | Authors: | Qiao, Z, Gao, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and computational examination of theArabidopsisprofilin-Poly-P complex reveals mechanistic details in profilin-regulated actin assembly.
J.Biol.Chem., 294, 2019
|
|
8Y5H
 
 | |
8Y5G
 
 | |
8Y5F
 
 | | Cryo-EM structure of E.coli spermidine transporter PotABC | | Descriptor: | Spermidine/putrescine import ATP-binding protein PotA, Spermidine/putrescine transport system permease protein PotB, Spermidine/putrescine transport system permease protein PotC | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural insights into polyamine spermidine uptake by the ABC transporter PotD-PotABC.
Sci Adv, 10, 2024
|
|
8Y5I
 
 | |
8ZX1
 
 | | Cryo-EM structure of E.coli spermidine transporter PotABC in nanodisc | | Descriptor: | Spermidine/putrescine ABC transporter membrane protein, Spermidine/putrescine import ATP-binding protein PotA, Spermidine/putrescine transport system permease protein PotC | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2024-06-13 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into polyamine spermidine uptake by the ABC transporter PotD-PotABC.
Sci Adv, 10, 2024
|
|
6IQF
 
 | | crystal structure of Arabidopsis thaliana Profilin 3 | | Descriptor: | PRF3 | | Authors: | Qiao, Z, Gao, Y. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.457 Å) | | Cite: | Structural and computational examination of theArabidopsisprofilin-Poly-P complex reveals mechanistic details in profilin-regulated actin assembly.
J.Biol.Chem., 294, 2019
|
|
6IQK
 
 | | crystal structure of Arabidopsis thaliana Profilin 3 | | Descriptor: | AtPRF3, Profilin-5 | | Authors: | Qiao, Z, Gao, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and computational examination of theArabidopsisprofilin-Poly-P complex reveals mechanistic details in profilin-regulated actin assembly.
J.Biol.Chem., 294, 2019
|
|
7CK1
 
 | | Crystal structure of arabidopsis CESA3 catalytic domain | | Descriptor: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming], MANGANESE (II) ION | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2020-07-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK3
 
 | | Crystal structure of Arabidopsis CESA3 catalytic domain | | Descriptor: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming] | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2020-07-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK2
 
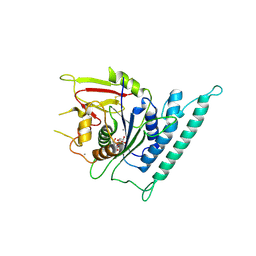 | | Crystal structure of Arabidopsis CESA3 catalytic domain with UDP-Glucose | | Descriptor: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming], MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2020-07-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WI3
 
 | | Cryo-EM structure of E.Coli FtsH-HflkC AAA protease complex | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Modulator of FtsH protease HflK | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
7WI4
 
 | | Cryo-EM structure of E.Coli FtsH protease cytosolic domains | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
8Z5G
 
 | |
4RF9
 
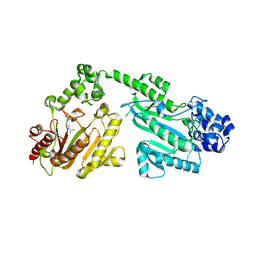 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with L-arginine and ATPgS | | Descriptor: | ACETATE ION, ARGININE, Arginine kinase, ... | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RF6
 
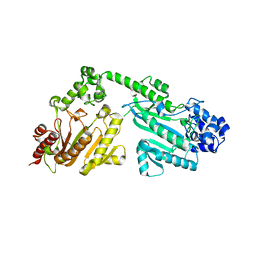 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas | | Descriptor: | Arginine kinase | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RF8
 
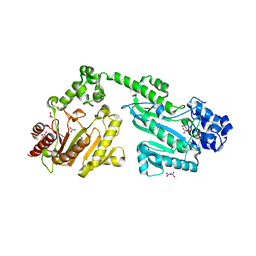 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, ... | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RF7
 
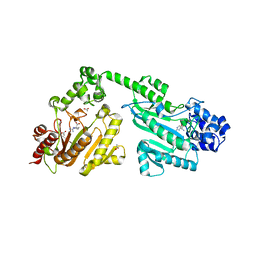 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with substrate L-arginine | | Descriptor: | ACETATE ION, ARGININE, Arginine kinase | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7RCU
 
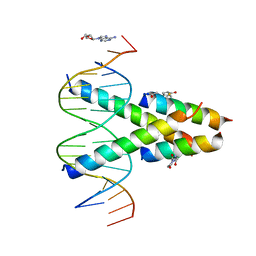 | | Synthetic Max homodimer mimic in complex with DNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2-(2,5-dioxopyrrolidin-1-yl)acetamide, ACETAMIDE, ... | | Authors: | Speltz, T, Qiao, Z, Shangguan, S, Fanning, S, Greene, J, Moellering, R. | | Deposit date: | 2021-07-08 | | Release date: | 2022-09-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains.
Nat.Biotechnol., 41, 2023
|
|
6IUY
 
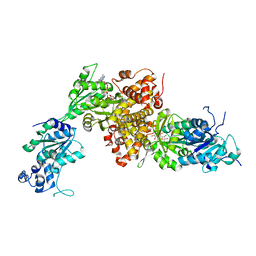 | | Structure of DsGPDH of Dunaliella salina | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Glycerol-3-phosphate dehydrogenase [NAD(+)], ... | | Authors: | He, Q, Toh, J.D, Ero, R, Qiao, Z, Kumar, V, Gao, Y.G. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The unusual di-domain structure of Dunaliella salina glycerol-3-phosphate dehydrogenase enables direct conversion of dihydroxyacetone phosphate to glycerol.
Plant J., 102, 2020
|
|
7WQ5
 
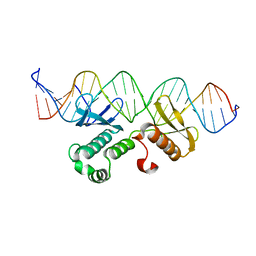 | | Crystal structure of Arabidopsis transcriptional factor WRINKLED1 with dsDNA | | Descriptor: | AMMONIUM ION, DNA (5'-D(P*GP*TP*GP*GP*AP*CP*GP*AP*TP*GP*AP*AP*AP*CP*CP*GP*AP*GP*GP*AP*AP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*TP*TP*CP*CP*TP*CP*GP*GP*TP*TP*TP*CP*AP*TP*CP*GP*TP*CP*CP*AP*C)-3'), ... | | Authors: | Zhu, Q, Gao, Y.G. | | Deposit date: | 2022-01-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of the key regulator WRINKLED1 in plant oil biosynthesis.
Sci Adv, 8, 2022
|
|
2PV6
 
 | | HIV-1 gp41 Membrane Proximal Ectodomain Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein | | Authors: | Sun, Z.-Y.J, Oh, K.J, Kim, M, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-05-09 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane
Immunity, 28, 2008
|
|
5EQJ
 
 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase from Saccharomyces cerevisiae | | Descriptor: | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
5ERG
 
 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase TRM6-TRM61 in complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
