5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | 分子名称: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | 著者 | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | 登録日 | 2016-11-20 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | 分子名称: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | 著者 | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2011-05-02 | | 公開日 | 2011-08-10 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4XAU
 
 | | Crystal structure of AtS13 from Actinomadura melliaura | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative aminotransferase | | 著者 | Wang, F, Singh, S, Xu, W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-12-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.0012 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
4XRR
 
 | | Crystal structure of cals8 from micromonospora echinospora (P294S mutant) | | 分子名称: | CalS8, GLYCEROL | | 著者 | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-01-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
J. Biol. Chem., 290, 2015
|
|
4XT0
 
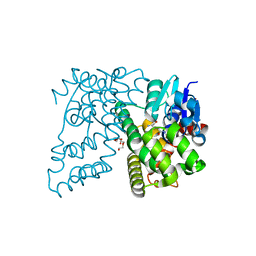 | | Crystal Structure of Beta-etherase LigF from Sphingobium sp. strain SYK-6 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, PENTAETHYLENE GLYCOL, ... | | 著者 | Helmich, K.E, Bingman, C.A, Donohue, T.J, Phillips Jr, G.N. | | 登録日 | 2015-01-22 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of beta-Aryl Ether Bonds in Lignin.
J.Biol.Chem., 291, 2016
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | 分子名称: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | 著者 | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | 登録日 | 2016-11-20 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
3KEV
 
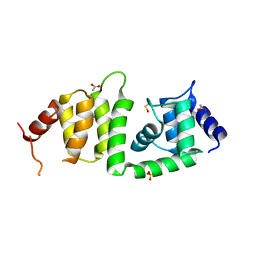 | | X-ray crystal structure of a DCUN1 domain-containing protein from Galdieria sulfuraria | | 分子名称: | ACETATE ION, Galieria sulfuraria DCUN1 domain-containing protein, SULFATE ION | | 著者 | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2009-10-26 | | 公開日 | 2009-12-01 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural architecture of Galdieria sulphuraria DCN1L.
Proteins, 79, 2011
|
|
5INJ
 
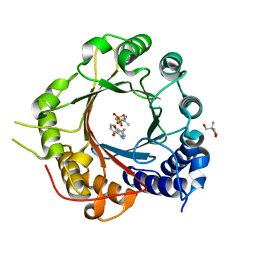 | | Crystal Structure of Prenyltransferase PriB Ternary Complex with L-Tryptophan and Dimethylallyl thiolodiphosphate (DMSPP) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Prenyltransferase, ... | | 著者 | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2016-03-07 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5HOQ
 
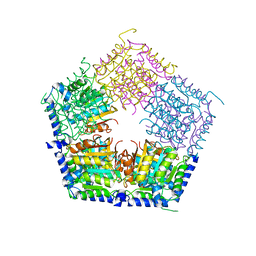 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | 分子名称: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | 著者 | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | 登録日 | 2016-01-19 | | 公開日 | 2016-03-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.793 Å) | | 主引用文献 | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
5K9M
 
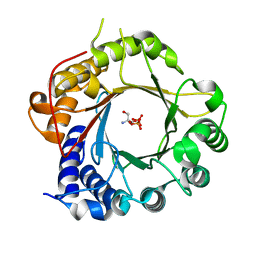 | | Crystal Structure of PriB Binary Complex with Product Diphosphate | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYROPHOSPHATE 2-, PriB Prenyltransferase | | 著者 | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2016-06-01 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5JXM
 
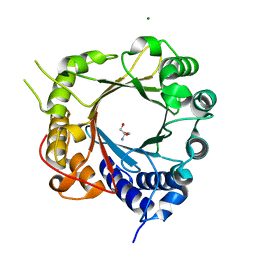 | | Crystal Structure of Prenyltransferase PriB Apo Form | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PriB | | 著者 | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2016-05-13 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
6BMG
 
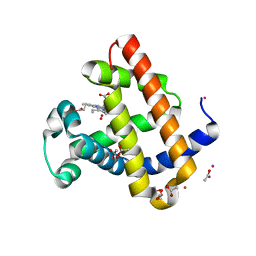 | | Structure of Recombinant Dwarf Sperm Whale Myoglobin (Oxy) | | 分子名称: | ACETATE ION, CADMIUM ION, Myoglobin, ... | | 著者 | Samuel, P.P, Miller, M.D, Xu, W, Alvarado, S, Phillips Jr, G.N, Olson, J.S. | | 登録日 | 2017-11-14 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structure of Recombinant Dwarf Sperm Whale Myoglobin (Oxy)
To Be Published
|
|
4W79
 
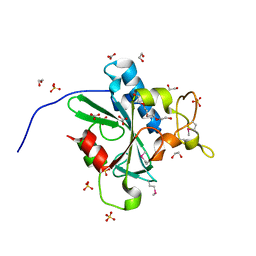 | | Crystal Structure of Human Protein N-terminal Glutamine Amidohydrolase | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, Protein N-terminal glutamine amidohydrolase, ... | | 著者 | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2014-08-21 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway.
Plos One, 9, 2014
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | 分子名称: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | 著者 | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-10-03 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2A3L
 
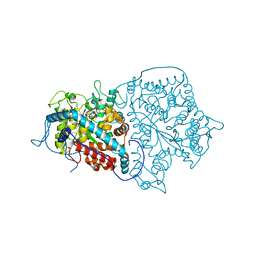 | | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate | | 分子名称: | AMP deaminase, COFORMYCIN 5'-PHOSPHATE, PHOSPHATE ION, ... | | 著者 | Han, B.W, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2005-06-25 | | 公開日 | 2005-07-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.34 Å) | | 主引用文献 | Membrane association, mechanism of action, and structure of Arabidopsis embryonic factor 1 (FAC1).
J.Biol.Chem., 281, 2006
|
|
3RI7
 
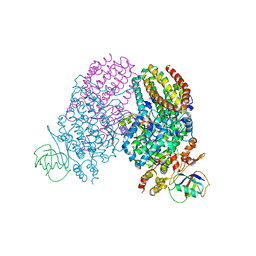 | | Toluene 4 monooxygenase HD Mutant G103L | | 分子名称: | ACETATE ION, FE (III) ION, Toluene-4-monooxygenase system protein A, ... | | 著者 | Bailey, L.J, Elsen, N.L, Fox, B.G. | | 登録日 | 2011-04-13 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
6AX6
 
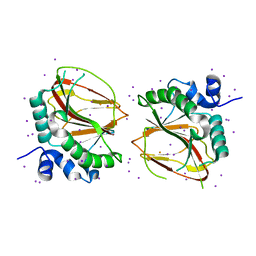 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | 分子名称: | FE (II) ION, IODIDE ION, Procollagen lysyl hydroxylase and glycosyltransferase | | 著者 | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | 登録日 | 2017-09-06 | | 公開日 | 2018-02-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.241 Å) | | 主引用文献 | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
6AX7
 
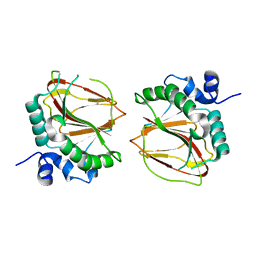 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | 分子名称: | FE (II) ION, Procollagen lysyl hydroxylase and glycosyltransferase | | 著者 | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | 登録日 | 2017-09-06 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | 分子名称: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | 著者 | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-07 | | 公開日 | 2014-05-07 | | 最終更新日 | 2015-02-11 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4PYT
 
 | | Crystal structure of a MurB family EP-UDP-N-acetylglucosamine reductase | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | 著者 | Cao, H, Franz, L, Sen, S, Bingman, C.A, Auldridge, M, Steinmetz, E, Mead, D, Phillips Jr, G.N. | | 登録日 | 2014-03-27 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.853 Å) | | 主引用文献 | LucY: A Versatile New Fluorescent Reporter Protein.
Plos One, 10, 2015
|
|
2A13
 
 | | X-ray structure of protein from Arabidopsis thaliana AT1G79260 | | 分子名称: | At1g79260 | | 著者 | Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2005-06-17 | | 公開日 | 2005-07-12 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
Proteins, 78, 2010
|
|
4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-04-10 | | 公開日 | 2014-05-07 | | 最終更新日 | 2017-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | 分子名称: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-08-29 | | 公開日 | 2014-10-01 | | 最終更新日 | 2016-11-02 | | 実験手法 | X-RAY DIFFRACTION (1.659 Å) | | 主引用文献 | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | 分子名称: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | 著者 | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-07-25 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.902 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | 著者 | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-05-27 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.4289 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
