2L8R
 
 | |
2LGR
 
 | |
5I7Z
 
 | | Crystal structure of a Par-6 PDZ-Crumbs 3 C-terminal peptide complex | | Descriptor: | Crb-3, DI(HYDROXYETHYL)ETHER, LD29223p | | Authors: | Whitney, D.S, Peterson, F.C, Prehoda, K.E, Volkman, B.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Binding of Crumbs to the Par-6 CRIB-PDZ Module Is Regulated by Cdc42.
Biochemistry, 55, 2016
|
|
8EY0
 
 | | Structure of an orthogonal PYR1*:HAB1* chemical-induced dimerization module in complex with mandipropamid | | Descriptor: | (2S)-2-(4-chlorophenyl)-N-{2-[3-methoxy-4-(prop-2-yn-1-yloxy)phenyl]ethyl}-2-(prop-2-yn-1-yloxy)ethanamide, Abscisic acid receptor PYR1, GLYCEROL, ... | | Authors: | Park, S.-Y, Volkman, B.F, Cutler, S.R, Peterson, F.C. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity.
Nat.Chem.Biol., 20, 2024
|
|
5KTF
 
 | | Structure of the C-terminal transmembrane domain of scavenger receptor BI (SR-BI) | | Descriptor: | Scavenger receptor class B member 1 | | Authors: | Chadwick, A.C, Peterson, F.C, Volkman, B.F, Sahoo, D. | | Deposit date: | 2016-07-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the C-Terminal Transmembrane Domain of the HDL Receptor, SR-BI, and a Functionally Relevant Leucine Zipper Motif.
Structure, 25, 2017
|
|
7JH1
 
 | |
4A1M
 
 | | NMR Structure of protoporphyrin-IX bound murine p22HBP | | Descriptor: | HEME-BINDING PROTEIN 1 | | Authors: | Goodfellow, B.J, Dias, J.S, Macedo, A.L, Ferreira, G.C, Peterson, F.C, Volkman, B.F, Duarte, I.C.N. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of Protoporphyrin-Ix Bound Murine P22Hbp
To be Published
|
|
6CWS
 
 | |
2GOV
 
 | | Solution structure of Murine p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Volkman, B.F, Dias, J.S, Goodfellow, B.J, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-14 | | Release date: | 2006-05-09 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The First Structure from the SOUL/HBP Family of Heme-binding Proteins, Murine P22HBP.
J.Biol.Chem., 281, 2006
|
|
1MKE
 
 | | Structure of the N-WASP EVH1 Domain-WIP complex | | Descriptor: | Fusion protein consisting of Wiskott-Aldrich syndrome protein interacting protein (WIP), GSGSG linker, and Neural Wiskott-Aldrich syndrome protein (N-WASP) | | Authors: | Volkman, B.F, Prehoda, K.E, Scott, J.A, Peterson, F.C, Lim, W.A. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-WASP EVH1 Domain-WIP Complex. Insight into the
Molecular Basis of Wiskott-Aldrich Syndrome.
Cell(Cambridge,Mass.), 111, 2002
|
|
1Q53
 
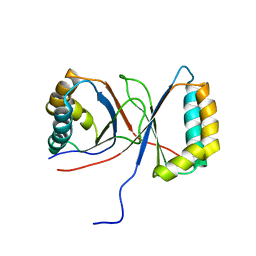 | |
5UR7
 
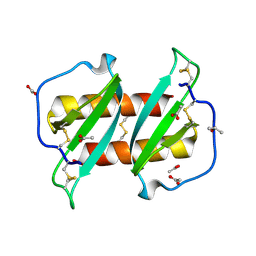 | |
5US5
 
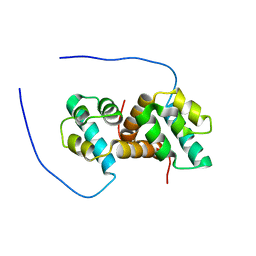 | | Solution structure of the IreB homodimer | | Descriptor: | UPF0297 protein EF_1202 | | Authors: | Lytle, B.L, Peterson, F.C, Volkman, B.F, Kristich, C.J, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2017-02-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dimerization of IreB, a Negative Regulator of Cephalosporin Resistance in Enterococcus faecalis.
J. Mol. Biol., 429, 2017
|
|
2G0Q
 
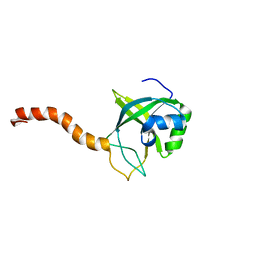 | | Solution structure of At5g39720.1 from Arabidopsis thaliana | | Descriptor: | AT5G39720.1 protein | | Authors: | Volkman, B.F, Peterson, F.C, Lytle, B.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-13 | | Release date: | 2006-02-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Arabidopsis thaliana protein At5g39720.1, a member of the AIG2-like protein family.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2GOW
 
 | | Solution structure of BC059385 from Homo sapiens | | Descriptor: | Ubiquitin-like protein 3 | | Authors: | Volkman, B.F, de la Cruz, N.B, Lytle, B.L, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-14 | | Release date: | 2006-04-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a membrane-anchored ubiquitin-fold (MUB) protein from Homo sapiens.
Protein Sci., 16, 2007
|
|
2HDM
 
 | |
2I9Y
 
 | | Solution structure of Arabidopsis thaliana protein At1g70830, a member of the major latex protein family | | Descriptor: | major latex protein-like protein 28 or MLP-like protein 28 | | Authors: | Volkman, B.F, de la Cruz, N.B, Lytle, B.L, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-09-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structures of two Arabidopsis thaliana major latex proteins represent novel helix-grip folds.
Proteins, 76, 2009
|
|
1ZR9
 
 | |
3K41
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M bound to Man-6-P | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3K43
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3K42
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-dependent mannose-6-phosphate receptor, SN-GLYCEROL-1-PHOSPHATE, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3LHR
 
 | | Crystal structure of the SCAN domain from Human ZNF24 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ETHYL MERCURY ION, ... | | Authors: | Volkman, B.F, Peterson, F.C, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SCAN domain from Human ZNF24
To be Published
|
|
7LP5
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin,E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LP4
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
1RJJ
 
 | | Solution structure of a homodimeric hypothetical protein, At5g22580, a structural genomics target from Arabidopsis thaliana | | Descriptor: | expressed protein | | Authors: | Cornilescu, G, Cornilescu, C.C, Zhao, Q, Frederick, R.O, Peterson, F.C, Thao, S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a homodimeric hypothetical protein, at5g22580, a structural genomics target from Arabidopsis thaliana.
J.Biomol.Nmr, 29, 2004
|
|
