3PNC
 
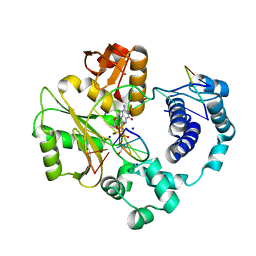 | | Ternary crystal structure of a polymerase lambda variant with a GT mispair at the primer terminus and sodium at catalytic metal site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*CP*AP*GP*TP*AP*G)-3', ... | | Authors: | Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2010-11-18 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Replication infidelity via a mismatch with Watson-Crick geometry.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PMN
 
 | | ternary crystal structure of polymerase lambda variant with a GT mispair at the primer terminus with Mn2+ in the active site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, 5'-D(*CP*AP*GP*TP*AP*G)-3', 5'-D(*CP*GP*GP*CP*CP*TP*TP*AP*CP*TP*G)-3', ... | | Authors: | Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Replication infidelity via a mismatch with Watson-Crick geometry.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4QH0
 
 | | Crystal structure of NucA from Streptococcus agalactiae with magnesium ion bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-entry nuclease (Competence-specific nuclease), ... | | Authors: | Moon, A.F, Gaudu, P, Pedersen, L.C. | | Deposit date: | 2014-05-26 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the virulence factor nuclease A from Streptococcus agalactiae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1VKJ
 
 | | Crystal structure of heparan sulfate 3-O-sulfotransferase isoform 1 in the presence of PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SULFATE ION, heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | | Authors: | Thorp, S, Lee, K.A, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1
J.Biol.Chem., 279, 2004
|
|
1QXM
 
 | | Crystal structure of a hemagglutinin component (HA1) from type C Clostridium botulinum | | Descriptor: | 1,2-ETHANEDIOL, HA1 | | Authors: | Inoue, K, Sobhany, M, Transue, T.R, Oguma, K, Pedersen, L.C, Negishi, M. | | Deposit date: | 2003-09-08 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis by X-ray crystallography and calorimetry of a haemagglutinin component (HA1) of the progenitor toxin from Clostridium botulinum.
Microbiology, 149, 2003
|
|
3C5G
 
 | | Structure of a ternary complex of the R517K Pol lambda mutant | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*DCP*DAP*DGP*DTP*DAP*(2DT))-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Foley, M.C, Pedersen, L.C, Schlick, T, Kunkel, T.A. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-29 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda.
Embo Rep., 9, 2008
|
|
3C5F
 
 | | Structure of a binary complex of the R517A Pol lambda mutant | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DTP*DAP*DC)-3'), DNA (5'-D(*DCP*DGP*DGP*DCP*DCP*DGP*DTP*DAP*DCP*DTP*DG)-3'), DNA (5'-D(P*DGP*DCP*DCP*DG)-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Foley, M.C, Pedersen, L.C, Schlick, T, Kunkel, T.A. | | Deposit date: | 2008-01-31 | | Release date: | 2008-09-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda.
Embo Rep., 9, 2008
|
|
3CU0
 
 | | human beta 1,3-glucuronyltransferase I (GlcAT-I) in complex with UDP and GAL-GAL(6-SO4)-XYL(2-PO4)-O-SER | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 3, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Tone, Y, Pedersen, L.C, Yamamoto, T, Kitagawa, H, Nishihara-Shimizu, J, Tamura, J, Negishi, M, Sugahara, K. | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-o-phosphorylation of xylose and 6-o-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis.
J.Biol.Chem., 283, 2008
|
|
3CQ8
 
 | | Ternary complex of the L415F mutant RB69 exo(-)polymerase | | Descriptor: | CALCIUM ION, DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*DCP*DC)-3'), ... | | Authors: | Zhong, X, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2008-04-02 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of a replicative DNA polymerase mutant with reduced fidelity and increased translesion synthesis capacity.
Nucleic Acids Res., 36, 2008
|
|
3C2K
 
 | | DNA POLYMERASE BETA with a gapped DNA substrate and DUMPNPP with Manganese in the active site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
3DM0
 
 | | Maltose Binding Protein fusion with RACK1 from A. thaliana | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein fused with RACK1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ullah, H, Scappini, E.L, Moon, A.F, Williams, L.V, Armstrong, D.L, Pedersen, L.C. | | Deposit date: | 2008-06-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana.
Protein Sci., 17, 2008
|
|
3C2M
 
 | | Ternary complex of DNA POLYMERASE BETA with a G:dAPCPP mismatch in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DGP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
1T8T
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CITRIC ACID, heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1 | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
1T8U
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP and tetrasaccharide substrate | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
1ZJN
 
 | | Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2005-04-29 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate.
STRUCTURE, 13, 2005
|
|
3C2L
 
 | | Ternary complex of DNA POLYMERASE BETA with a C:DAPCPP mismatch in the active site | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DCP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DTP*DGP*DAP*DTP*DGP*DCP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
1ZM8
 
 | | Apo Crystal structure of Nuclease A from Anabaena sp. | | Descriptor: | MANGANESE (II) ION, Nuclease, SODIUM ION, ... | | Authors: | Ghosh, M, Meiss, G, Pingoud, A, London, R.E, Pedersen, L.C. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Mechanism of Nuclease A, a beta beta alpha Metal Nuclease from Anabaena.
J.Biol.Chem., 280, 2005
|
|
3F5F
 
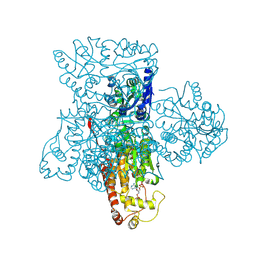 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | Authors: | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3UAN
 
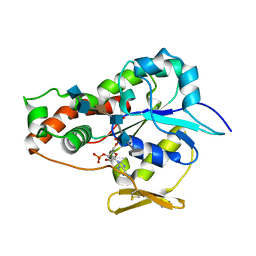 | | Crystal structure of 3-O-sulfotransferase (3-OST-1) with bound PAP and heptasaccharide substrate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Moon, A.F, Xu, Y, Woody, S.M, Krahn, J.M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7M0E
 
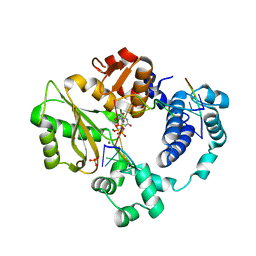 | | Pre-catalytic synaptic complex of DNA Polymerase Lambda with gapped DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M09
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with blunt-ended DSB substrate and incoming dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0A
 
 | | Incomplete in crystallo incorporation by DNA Polymerase Lambda bound to blunt-ended DSB substrate and incoming dTTP | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0D
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound complementary DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0B
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound mismatched DSB and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0C
 
 | | Post-catalytic nicked complex of DNA Polymerase Lambda with bound mismatched DSB substrate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Comprehensive structural survey of DNA double-strand break synapsis by DNA Polymerase Lambda
Not Published
|
|
