3ZVM
 
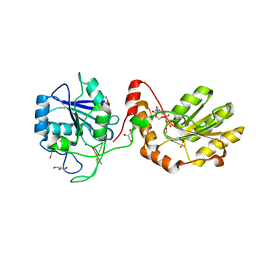 | | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*GP*TP*CP*AP*CP)-3', ACETATE ION, ... | | Authors: | Garces, F, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase.
Mol. Cell, 44, 2011
|
|
4BU1
 
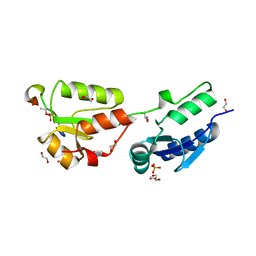 | | Crystal structure of Rad4 BRCT1,2 in complex with a Crb2 phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, DNA REPAIR PROTEIN RHP9, GLYCEROL, ... | | Authors: | Qu, M, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-06-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
4BMC
 
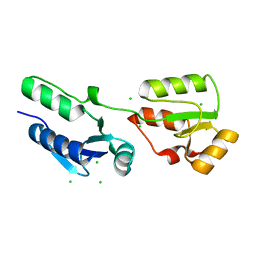 | | Crystal structure of s.pombe Rad4 BRCT1,2 | | Descriptor: | CHLORIDE ION, S-M CHECKPOINT CONTROL PROTEIN RAD4 | | Authors: | Meng, Q, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
4BMD
 
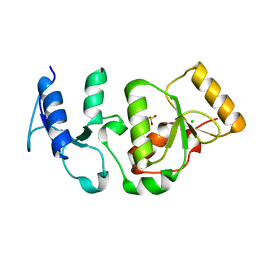 | | Crystal structure of S.pombe Rad4 BRCT3,4 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, S-M CHECKPOINT CONTROL PROTEIN RAD4 | | Authors: | Meng, Q, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
3ZVN
 
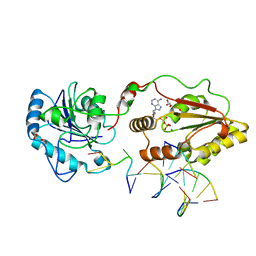 | | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase | | Descriptor: | 5'-D(*GP*TP*CP*AP*CP)-3', ADENOSINE-5'-DIPHOSPHATE, BIFUNCTIONAL POLYNUCLEOTIDE PHOSPHATASE/KINASE, ... | | Authors: | Garces, F, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase.
Mol. Cell, 44, 2011
|
|
4BU0
 
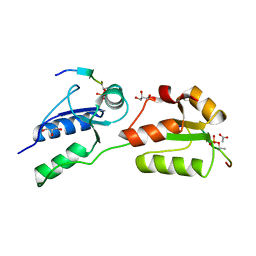 | | Crystal structure of Rad4 BRCT1,2 in complex with a Crb2 phosphopeptide | | Descriptor: | ACETATE ION, DNA REPAIR PROTEIN RHP9, GLYCEROL, ... | | Authors: | Qu, M, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-06-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
1GEC
 
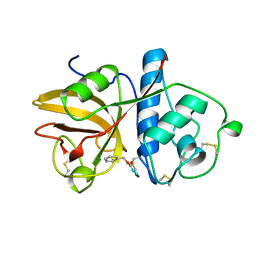 | | GLYCYL ENDOPEPTIDASE-COMPLEX WITH BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE COVALENTLY BOUND TO CYSTEINE 25 | | Descriptor: | BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE INHIBITOR, GLYCYL ENDOPEPTIDASE | | Authors: | Ohara, B.P, Hemmings, A.M, Buttle, D.J, Pearl, L.H. | | Deposit date: | 1995-05-25 | | Release date: | 1995-12-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glycyl endopeptidase from Carica papaya: a cysteine endopeptidase of unusual substrate specificity.
Biochemistry, 34, 1995
|
|
1GML
 
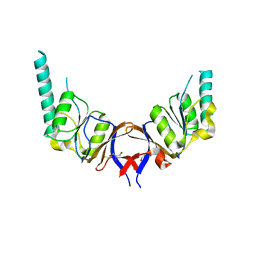 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | Descriptor: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-09-17 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GN1
 
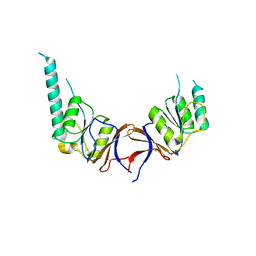 | | crystal structure of the mouse CCT gamma apical domain (monoclinic) | | Descriptor: | CALCIUM ION, CCT-GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-10-01 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1H0C
 
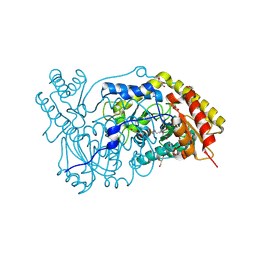 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
1BGQ
 
 | |
1AH6
 
 | |
1AH8
 
 | |
1HK7
 
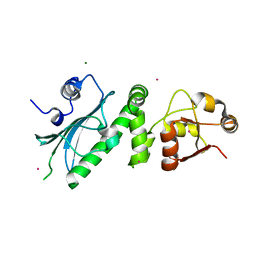 | | Middle Domain of HSP90 | | Descriptor: | CADMIUM ION, HEAT SHOCK PROTEIN HSP82, MAGNESIUM ION | | Authors: | Meyer, P, Prodromou, C, Roe, S.M, Pearl, L.H. | | Deposit date: | 2003-03-06 | | Release date: | 2004-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of the Middle Segment of Hsp90. Implications for ATP Hydrolysis and Client Protein and Cochaperone Interactions
Mol.Cell, 11, 2003
|
|
1KCF
 
 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
1KO9
 
 | | Native Structure of the Human 8-oxoguanine DNA Glycosylase hOGG1 | | Descriptor: | 8-oxoguanine DNA glycosylase, SULFATE ION | | Authors: | Bjoras, M, Seeberg, E, Luna, L, Pearl, L.H, Barrett, T.E. | | Deposit date: | 2001-12-20 | | Release date: | 2002-01-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Reciprocal "flipping" underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase.
J.Mol.Biol., 317, 2002
|
|
1A4H
 
 | |
7AUD
 
 | |
7AUC
 
 | |
4ER4
 
 | | HIGH-RESOLUTION X-RAY ANALYSES OF RENIN INHIBITOR-ASPARTIC PROTEINASE COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, H-142 | | Authors: | Foundling, S.I, Watson, F.E, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Nature, 327, 1987
|
|
7OLE
 
 | | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2, ... | | Authors: | Pal, M, Llorca, O, Pearl, L. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone.
Cell Rep, 36, 2021
|
|
5MGX
 
 | |
4CV4
 
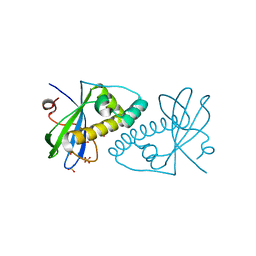 | | PIH N-terminal domain | | Descriptor: | COBALT (II) ION, PIH1 DOMAIN-CONTAINING PROTEIN 1, SULFATE ION | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-03-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CSE
 
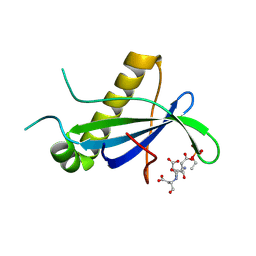 | | PIH N-terminal domain | | Descriptor: | PIH1 DOMAIN-CONTAINING PROTEIN 1, TELOMERE LENGTH REGULATION PROTEIN TEL2 HOMOLOG | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4U6A
 
 | |
