4DFD
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from bacteroides thetaiotaomicron, magnesium complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative haloacid dehalogenase-like hydrolase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
1TE5
 
 | |
4LFE
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, partially liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl diphosphate synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
4LFG
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, fully liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl Diphosphate Synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
4LOB
 
 | | Crystal structure of polyprenyl diphosphate synthase A1S_2732 (Target EFI-509223) from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, GLYCEROL, Polyprenyl synthetase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of polyprenyl diphosphate synthase from Acinetobacter baumannii
To be Published
|
|
3SJN
 
 | | Crystal structure of enolase Spea_3858 (target EFI-500646) from Shewanella pealeana with magnesium bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Enolase Spea_3858 from Shewanella Pealeana
To be Published
|
|
3K9D
 
 | | CRYSTAL STRUCTURE OF PROBABLE ALDEHYDE DEHYDROGENASE FROM Listeria monocytogenes EGD-e | | Descriptor: | ALDEHYDE DEHYDROGENASE, CHLORIDE ION, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase from Listeria Monocytogenes
To be Published
|
|
3KEW
 
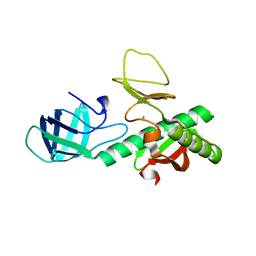 | | Crystal structure of probable alanyl-trna-synthase from Clostridium perfringens | | Descriptor: | DHHA1 domain protein, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alanyl-trna-synthase from Clostridium perfringens
To be Published
|
|
1S7J
 
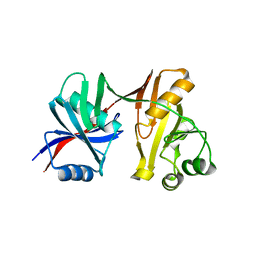 | |
2F3M
 
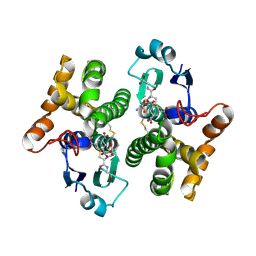 | | Structure of human GLUTATHIONE S-TRANSFERASE M1A-1A complexed with 1-(S-(GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXADIENATE ANION | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
2QUP
 
 | | Crystal structure of uncharacterized protein BH1478 from Bacillus halodurans | | Descriptor: | BH1478 protein, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Mckenzie, C, Bain, K.T, Smith, D, Ozyurt, S, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-06 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uncharacterized Protein Bh1478 from Bacillus Halodurans.
To be Published
|
|
2QH5
 
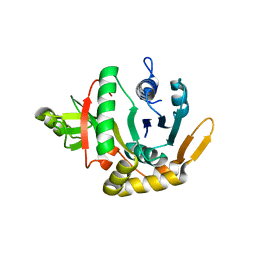 | | Crystal structure of mannose-6-phosphate isomerase from Helicobacter pylori | | Descriptor: | Mannose-6-phosphate isomerase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Iizuka, M, Maletic, M, Wasserman, S.R, Koss, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mannose-6-phosphate isomerase from Helicobacter pylori.
To be Published
|
|
2QSD
 
 | | Crystal structure of a protein Il1583 from Idiomarina loihiensis | | Descriptor: | GLYCEROL, Uncharacterized conserved protein | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Romero, R, Rutter, M, Koss, J, Mckenzie, C, Gheyi, T, Bain, K, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Protein Il1583 from Idiomarina loihiensis.
To be Published
|
|
2R1F
 
 | | Crystal structure of predicted aminodeoxychorismate lyase from Escherichia coli | | Descriptor: | CADMIUM ION, GLYCEROL, Predicted aminodeoxychorismate lyase, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Rutter, M, Lau, C, Maletic, M, Smith, D, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Predicted Aminodeoxychorismate Lyase from Escherichia coli.
To be Published
|
|
2QV5
 
 | | Crystal structure of uncharacterized protein ATU2773 from Agrobacterium tumefaciens C58 | | Descriptor: | GLYCEROL, Uncharacterized protein Atu2773 | | Authors: | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Dickey, M, Bain, K.T, Iizuka, M, Smith, D, Rodgers, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-07 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uncharacterized Protein Atu2773 from Agrobacterium Tumefaciens.
To be Published
|
|
2RDM
 
 | | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Yan, Q, Zhan, C, Toro, R, Meyer, A.J, Gilmore, M, Hu, S, Groshong, C, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419.
To be Published
|
|
2RDX
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Ozyurt, S, Gilmore, M, Lau, C, Maletic, M, Gheyi, T, Wasserman, S.R, Koss, J, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM.
To be Published
|
|
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3BY5
 
 | | Crystal structure of cobalamin biosynthesis protein chiG from Agrobacterium tumefaciens str. C58 | | Descriptor: | Cobalamin biosynthesis protein, SULFATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Rutter, M, Iizuka, M, Maletic, M, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of cobalamin biosynthesis protein from Agrobacterium tumefaciens str. C58.
To be Published
|
|
3C3M
 
 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3C6F
 
 | | Crystal structure of protein Bsu07140 from Bacillus subtilis | | Descriptor: | GLYCEROL, YetF protein | | Authors: | Patskovsky, Y, Min, T, Zhang, A, Adams, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein Bsu07140 from Bacillus subtilis.
To be Published
|
|
3BMA
 
 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | Descriptor: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
3BVC
 
 | | Crystal structure of uncharacterized protein Ism_01780 from Roseovarius nubinhibens ISM | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Uncharacterized protein Ism_01780 | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Rutter, M, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-06 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an uncharacterized protein Ism_01780 from Roseovarius nubinhibens.
To be Published
|
|
3BQ9
 
 | | Crystal structure of predicted nucleotide-binding protein from Idiomarina baltica OS145 | | Descriptor: | GLYCEROL, Predicted Rossmann fold nucleotide-binding domain-containing protein, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Dickey, M, Eberle, M, Koss, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Idiomarina baltica.
To be Published
|
|
3C9F
 
 | | Crystal structure of 5'-nucleotidase from Candida albicans SC5314 | | Descriptor: | 5'-nucleotidase, FORMIC ACID, SODIUM ION, ... | | Authors: | Patskovsky, Y, Romero, R, Gilmore, M, Eberle, M, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 5'-nucleotidase from Candida albicans.
To be Published
|
|
