8C5U
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG (IC8) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8C5S
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 7-mer RNA, pppGpGpUpApApApU (IC7) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8Q63
 
 | | Cryo-EM structure of IC8', a second state of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-08-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8AP1
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with two GTP molecules poised for de novo initiation (IC2) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATV
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 5-mer RNA, pppGpGpApApA (IC5) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATT
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 4-mer RNA, pppGpGpUpA (IC4) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATW
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 6-mer RNA, pppGpGpApApApU (IC6) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
6YMV
 
 | |
6YMW
 
 | |
4ZTU
 
 | | Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (25-MER), DNA (5'-D(P*AP*AP*GP*AP*CP*GP*AP*GP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*GP*TP*AP*C)-3'), ... | | Authors: | Szymanski, M.R, Kuznestov, V.B, Shumate, C.K, Meng, Q, Lee, Y.-S, Patel, G, Patel, S.S, Yin, Y.W. | | Deposit date: | 2015-05-15 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural basis for processivity and antiviral drug toxicity
in human mitochondrial DNA replicase
EMBO J., 34, 2015
|
|
5E3H
 
 | | Structural Basis for RNA Recognition and Activation of RIG-I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Jiang, F, Miller, M.T, Marcotrigiano, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of RNA recognition and activation by innate immune receptor RIG-I.
Nature, 479, 2011
|
|
5F98
 
 | | Crystal structure of RIG-I in complex with Cap-0 RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F9F
 
 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer blunt-end hairpin RNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M.T, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F9H
 
 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer 5' triphosphate hairpin RNA | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZTZ
 
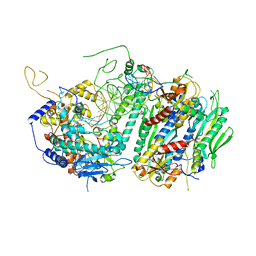 | |
2PL9
 
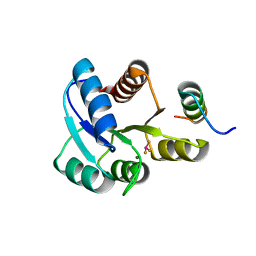 | |
2PMC
 
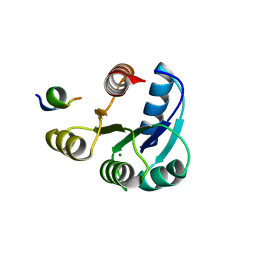 | |
