5WLS
 
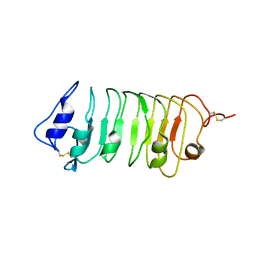 | | Crystal Structure of a Pollen Receptor Kinase 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pollen receptor-like kinase 3 | | Authors: | Xu, G, Chakraborty, S, Pan, H. | | Deposit date: | 2017-07-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | The Extracellular Domain of Pollen Receptor Kinase 3 is structurally similar to the SERK family of co-receptors.
Sci Rep, 8, 2018
|
|
4I11
 
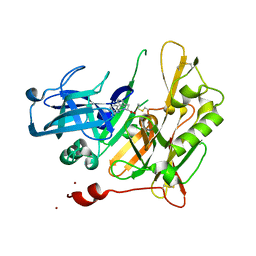 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates. | | Descriptor: | Beta-secretase 1, N-(3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)-L-phenylalanine, ZINC ION | | Authors: | Bowers, B, Xu, Y, Yuan, S, Probst, G.D, Hom, R.K, Chan, W, Konradi, A.W, Sham, H.L, Zhu, Y.L, Beroza, P, Pan, H, Brecht, E, Yao, N, Lougheed, J, Artis, D.R, Tam, D, Bova, M. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3OY1
 
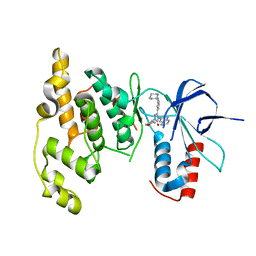 | | Highly Selective c-Jun N-Terminal Kinase (JNK) 2 and 3 Inhibitors with In Vitro CNS-like Pharmacokinetic Properties | | Descriptor: | 5-[2-(cyclohexylamino)pyridin-4-yl]-4-naphthalen-2-yl-2-(tetrahydro-2H-pyran-4-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Mitogen-activated protein kinase 10 | | Authors: | Probst, G.D, Bowers, S, Sealy, J.M, Truong, A, Neitz, J, Hom, R.K, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Quincy, D, Pan, H, Yao, N. | | Deposit date: | 2010-09-22 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Highly selective c-Jun N-terminal kinase (JNK) 2 and 3 inhibitors with in vitro CNS-like pharmacokinetic properties prevent neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PTG
 
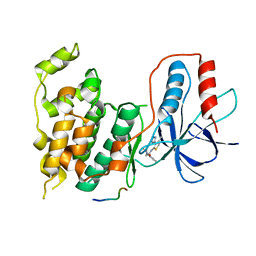 | | Design and Synthesis of a Novel, Orally Efficacious Tri-substituted Thiophene Based JNK Inhibitor | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 10, N-[4-methyl-3-(1H-1,2,4-triazol-5-yl)thiophen-2-yl]-2-(2-oxo-3,4-dihydroquinolin-1(2H)-yl)acetamide | | Authors: | Bowers, S, Truong, A.P, Neitz, J, Neitzel, M, Probst, G.D, Hom, R.K, Konradi, A.W, Sham, H.L, Toth, G, Pan, H, Yao, N, Artis, D.R, Brigham, E.F, Quinn, K.P, Sauer, J, Powell, K, Ruslim, L, Bard, F, Yednock, T.A, Griswold-Prenner, I. | | Deposit date: | 2010-12-02 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Design and synthesis of a novel, orally active, brain penetrant, tri-substituted thiophene based JNK inhibitor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6L30
 
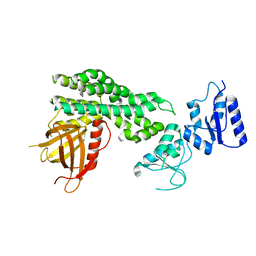 | | Crystal structure of the epithelial cell transforming 2 (ECT2) | | Descriptor: | Protein ECT2 | | Authors: | Chen, Z.C, Chen, M.R, Pan, H, Sun, L.F, Shi, P. | | Deposit date: | 2019-10-07 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and regulation of human epithelial cell transforming 2 protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4R1T
 
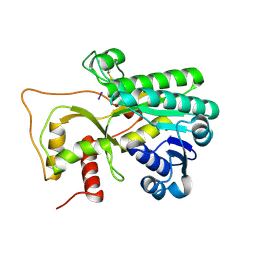 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | cinnamoyl CoA reductase, molecular iodine | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
4R1U
 
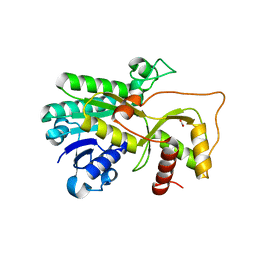 | | Crystal structure of Medicago truncatula cinnamoyl-CoA reductase | | Descriptor: | ACETATE ION, Cinnamoyl CoA reductase | | Authors: | Noel, J.P, Bomati, E.K, Louie, G.V, Bowman, M.E. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
4R1S
 
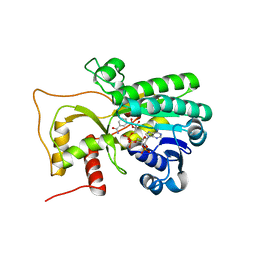 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, cinnamoyl CoA reductase | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
3MFE
 
 | |
3MKA
 
 | |
3MI0
 
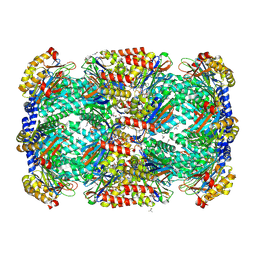 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome at 2.2 A | | Descriptor: | (2R,3S,4R)-2-[(S)-(1S)-cyclohex-2-en-1-yl(hydroxy)methyl]-4-ethyl-3-hydroxy-3-methyl-5-oxopyrrolidine-2-carbaldehyde, DIMETHYLFORMAMIDE, Proteasome subunit alpha, ... | | Authors: | Li, D, Li, H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome.
Embo J., 2010
|
|
8TWQ
 
 | |
8EPA
 
 | |
3HBF
 
 | | Structure of UGT78G1 complexed with myricetin and UDP | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
3HBJ
 
 | | Structure of UGT78G1 complexed with UDP | | Descriptor: | Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
1IJL
 
 | | Crystal structure of acidic phospholipase A2 from deinagkistrodon acutus | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, ZINC ION | | Authors: | Gu, L, Zhang, H, Song, S, Zhou, Y, Lin, Z. | | Deposit date: | 2001-04-27 | | Release date: | 2001-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an acidic phospholipase A2 from the venom of Deinagkistrodon acutus.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5U1M
 
 | |
2AXY
 
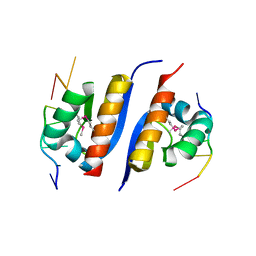 | | Crystal Structure of KH1 domain of human Poly(C)-binding protein-2 with C-rich strand of human telomeric DNA | | Descriptor: | C-rich strand of human telomeric dna, Poly(rC)-binding protein 2 | | Authors: | Du, Z, Lee, J.K, Tjhen, R.J, Li, S, Stroud, R.M, James, T.L. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the First KH Domain of Human Poly(C)-binding Protein-2 in Complex with a C-rich Strand of Human Telomeric DNA at 1.7 A
J.Biol.Chem., 280, 2005
|
|
3TLQ
 
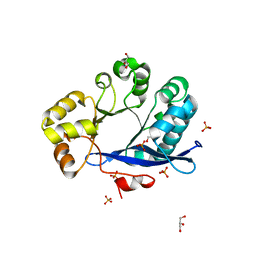 | | Crystal structure of EAL-like domain protein YdiV | | Descriptor: | GLYCEROL, PHOSPHATE ION, Regulatory protein YdiV | | Authors: | Li, B, Li, N, Wang, F, Guo, L, Liu, C, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
4ES4
 
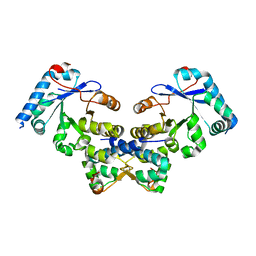 | | Crystal structure of YdiV and FlhD complex | | Descriptor: | Flagellar transcriptional regulator FlhD, Putative cyclic di-GMP regulator CdgR | | Authors: | Li, B, Gu, L. | | Deposit date: | 2012-04-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
8FQ7
 
 | | Nanobody with WIW inserted in CDR3 loop to Inhibit Growth of Alzheimer's Tau fibrils | | Descriptor: | GLYCEROL, Nanobody with WIW insert in CDR3 loop to target tau fibrils | | Authors: | Abskharon, R, Sawaya, M.R, Cascio, D.C, Eisenberg, D.S. | | Deposit date: | 2023-01-05 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of nanobodies that inhibit seeding of Alzheimer's patient-extracted tau fibrils.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4I0D
 
 | |
4I10
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates | | Descriptor: | 2-{(1S)-1-[(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)amino]-2-phenylethyl}pyrido[4,3-d]pyrimidin-4(1H)-one, Beta-secretase 1, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I12
 
 | | Design and synthesis of thiophene dihydroisoquinolins as novel BACE-1 inhibitors | | Descriptor: | 2-{(1S)-1-{[(1Z)-6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1(2H)-ylidene]amino}-2-[2-propyl-4-(1H-pyrazol-4-yl)thiophen-3-yl]ethyl}pyrimidin-4(5H)-one, Beta-secretase 1, SODIUM ION, ... | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Design and synthesis of thiophene dihydroisoquinolines as novel BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I0G
 
 | | Design and Synthesis of Thiophene Dihydroisoquinolins as Novel BACE-1 Inhibitors | | Descriptor: | 3-(4-bromothiophen-3-yl)-N-(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)-L-alanine, Beta-secretase 1, ZINC ION | | Authors: | Yao, N, Brecht, E. | | Deposit date: | 2012-11-16 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
