3P4D
 
 | | Alternatingly modified 2'Fluoro RNA octamer f/rC4G4 | | Descriptor: | 5'-R(*(CFZ)P*CP*(CFZ)P*CP*(GF2)P*GP*(GF2)P*G)-3' | | Authors: | Pallan, P.S, Greene, E.M, Jicman, P.A, Pandey, R.K, Manoharan, M, Rozners, E, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
5SWM
 
 | |
4DKZ
 
 | |
6B82
 
 | | Zebra Fish CYP-450 17A1 Mutant Abiraterone Complex | | Descriptor: | ACETATE ION, Abiraterone, CHLORIDE ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Inherent steroid 17 alpha ,20-lyase activity in defunct cytochrome P450 17A enzymes.
J. Biol. Chem., 293, 2018
|
|
6XHC
 
 | | Structure of glycinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]ethanoic acid, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHE
 
 | | Structure of beta-prolinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 5'-O-[(S)-[(3S)-3-carboxypyrrolidin-1-yl](hydroxy)phosphoryl]adenosine, ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHD
 
 | | Structure of Prolinyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-1-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]pyrrolidine-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHF
 
 | | Structure of Phenylalanyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]-3-phenyl-propanoic acid, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3I8D
 
 | |
7U38
 
 | | Pixantrone tethered DNA duplex | | Descriptor: | 1,2-ETHANEDIOL, Pixantrone AP conjugate-modified DNA | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-02-26 | | Release date: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Characterization of a Covalent Conjugate between Pixantrone and an Abasic Site in DNA
To Be Published
|
|
6VEM
 
 | | Structure of RNA octamer | | Descriptor: | COBALT HEXAMMINE(III), Modified Octamer RNA | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis, chirality-dependent conformational and biological properties of siRNAs containing 5'-(R)- and 5'-(S)-C-methyl-guanosine.
Nucleic Acids Res., 48, 2020
|
|
5DO4
 
 | | Thrombin-RNA aptamer complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Evoking picomolar binding in RNA by a single phosphorodithioate linkage.
Nucleic Acids Res., 44, 2016
|
|
8CTY
 
 | | 12-mer DNA structure of ExBIM bound to RNase-H | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformation and Pairing Properties of an O 6 -Methyl-2'-deoxyguanosine-Directed Benzimidazole Nucleoside Analog in Duplex DNA.
Chem.Res.Toxicol., 35, 2022
|
|
8CTZ
 
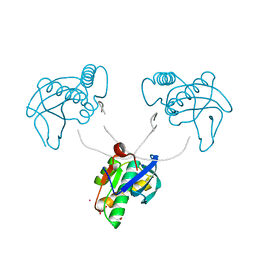 | | 12-mer DNA structure of ExBIM & O6Me-G bound to RNase-H | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*CP*GP*CP*(6OG)P*AP*AP*TP*TP*(OWR)P*GP*CP*G)-3'), ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Conformation and Pairing Properties of an O 6 -Methyl-2'-deoxyguanosine-Directed Benzimidazole Nucleoside Analog in Duplex DNA.
Chem.Res.Toxicol., 35, 2022
|
|
8CU0
 
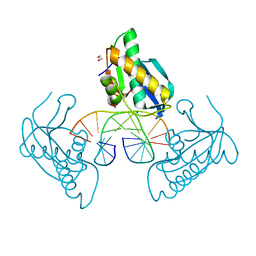 | | 12-mer DNA structure of ExBIM bound to RNaseH -modified DDD | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*(OWR)P*CP*CP*G)-3'), GLYCEROL, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Conformation and Pairing Properties of an O 6 -Methyl-2'-deoxyguanosine-Directed Benzimidazole Nucleoside Analog in Duplex DNA.
Chem.Res.Toxicol., 35, 2022
|
|
3EY2
 
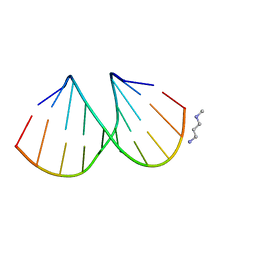 | |
3EY3
 
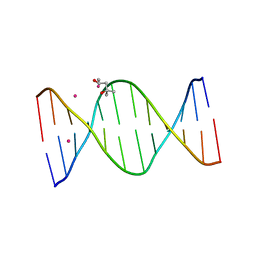 | |
3EY1
 
 | |
4O41
 
 | | Amide linked RNA | | Descriptor: | AMIDE LINKED RNA, STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs.
Nucleic Acids Res., 42, 2014
|
|
4OPK
 
 | | Bh-RNaseH:2'-SMe-DNA complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(USM)P*TP*CP*GP*CP*G)-3', GLYCEROL, Ribonuclease H | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2014-02-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Generating Crystallographic Models of DNA Dodecamers from Structures of RNase H:DNA Complexes.
Methods Mol.Biol., 1320
|
|
4OPJ
 
 | | Bh-RNaseH:tcdA-DNA complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*(TCY)P*TP*TP*CP*GP*CP*G)-3', GLYCEROL, Ribonuclease H | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2014-02-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Generating Crystallographic Models of DNA Dodecamers from Structures of RNase H:DNA Complexes.
Methods Mol.Biol., 1320
|
|
7JJF
 
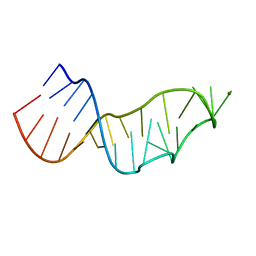 | | Sarcin-ricin loop with modified residue. | | Descriptor: | MAGNESIUM ION, RNA/DNA (27-mer) | | Authors: | Harp, J.M, Pallan, P.S, Egli, M. | | Deposit date: | 2020-07-25 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Incorporating a Thiophosphate Modification into a Common RNA Tetraloop Motif Causes an Unanticipated Stability Boost.
Biochemistry, 59, 2020
|
|
3P4A
 
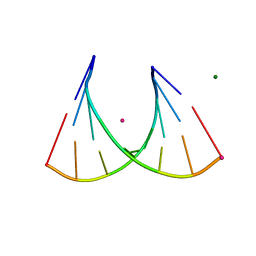 | | 2'Fluoro modified RNA octamer fA2U2 | | Descriptor: | 2'Fluoro modified RNA 8-MER, MAGNESIUM ION, STRONTIUM ION | | Authors: | Manoharan, M, Akinc, A, Pandey, R.K, Qin, J, Hadwiger, P, John, M, Mills, K, Charisse, K, Maier, M.A, Nechev, L, Greene, E.M, Pallan, P.S, Rozners, E, Rajeev, K.G, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
5VAJ
 
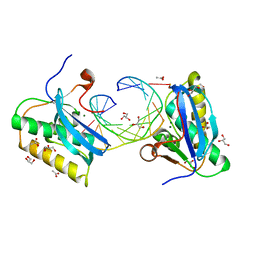 | | BhRNase H - amide-RNA/DNA complex | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), GLYCEROL, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs.
Nucleic Acids Res., 45, 2017
|
|
5VBU
 
 | |
