8H1I
 
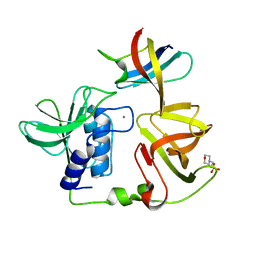 | | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cold shock-like protein CspC, ... | | Authors: | Padmanabhan, B, Gopinatha, K, Mandal, M, Saranya, G, Sudhagar, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C
To Be Published
|
|
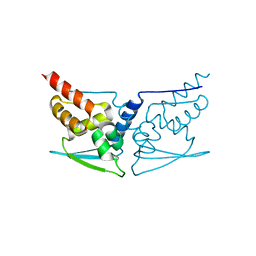
7EXI
 
 | |
4YZO
 
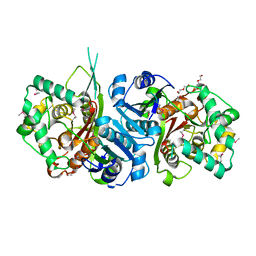 | | Crystal Structure Analysis of Thiolase-like protein, ST0096 from Sulfolobus Tokodaii | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Padmanabhan, B, Manjula, R, Yokoyama, S, Bessho, Y. | | Deposit date: | 2015-03-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Analysis of Thiolase-like protein, ST0096 from Sulfolobus Tokodaii
To Be Published
|
|
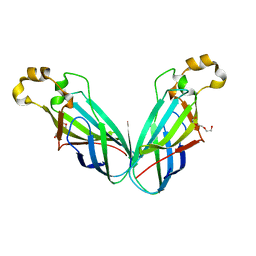
2DZE
 
 | |
1IMJ
 
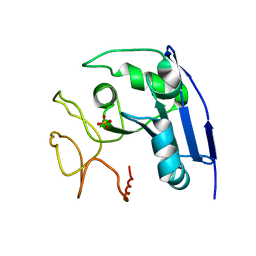 | |
1X2J
 
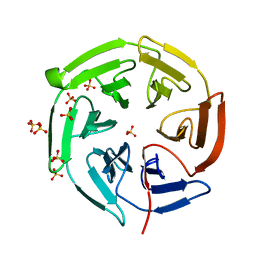 | | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Padmanabhan, B, Tong, K.I, Nakamura, Y, Ohta, T, Scharlock, M, Kobayashi, A, Ohtsuji, M, Kang, M.-I, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-25 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for defects of keap1 activity provoked by its point mutations in lung cancer
Mol.Cell, 21, 2006
|
|
1X2R
 
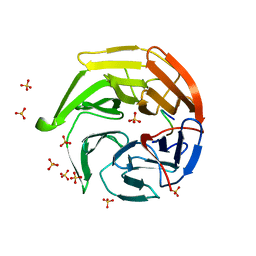 | | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | | Descriptor: | Kelch-like ECH-associated protein 1, Nuclear factor erythroid 2 related factor 2, SULFATE ION | | Authors: | Padmanabhan, B, Tong, K.I, Nakamura, Y, Ohta, T, Scharlock, M, Kobayashi, A, Ohtsuji, M, Kang, M.-I, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for defects of keap1 activity provoked by its point mutations in lung cancer
Mol.Cell, 21, 2006
|
|
1WG3
 
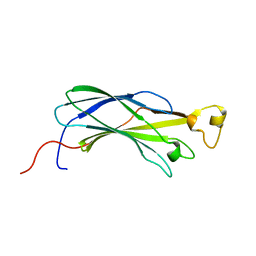 | | Structural analysis of yeast nucleosome-assembly factor CIA1p | | Descriptor: | Anti-silencing protein 1 | | Authors: | Padmanabhan, B, Kataoka, K, Umehara, T, Adachi, N, Yokoyama, S, Horikoshi, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Similarity between Histone Chaperone Cia1p/Asf1p and DNA-Binding Protein NF-{kappa}B
J.Biochem.(Tokyo), 138, 2005
|
|
1IXV
 
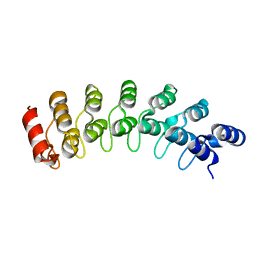 | | Crystal Structure Analysis of homolog of oncoprotein gankyrin, an interactor of Rb and CDK4/6 | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Padmanabhan, B, Adachi, N, Kataoka, K, Horikoshi, M. | | Deposit date: | 2002-07-09 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and CDK4/6
J.BIOL.CHEM., 279, 2004
|
|
1KP0
 
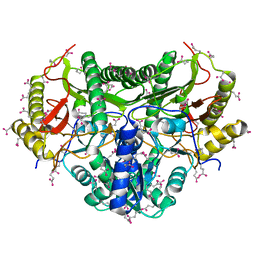 | |
2YXH
 
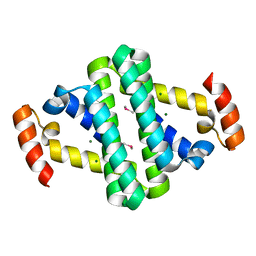 | |
2YXG
 
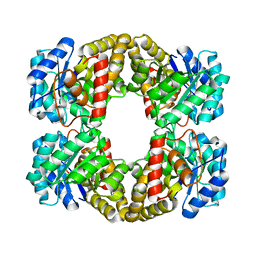 | |
2YXE
 
 | |
2ZG6
 
 | |
2ZGI
 
 | | Crystal Structure of Putative 4-amino-4-deoxychorismate lyase | | Descriptor: | DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE, Putative 4-amino-4-deoxychorismate lyase, ... | | Authors: | Padmanabhan, B, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-22 | | Release date: | 2008-07-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of putative 4-amino-4-deoxychorismate lyase from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2ZAD
 
 | |
2DWW
 
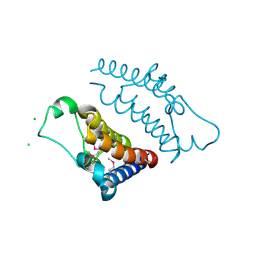 | |
2DVV
 
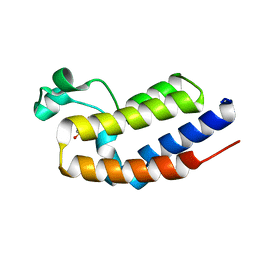 | |
2YXD
 
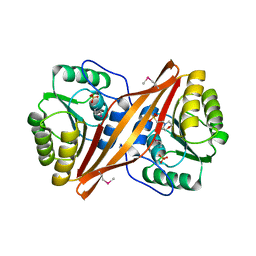 | |
2Z32
 
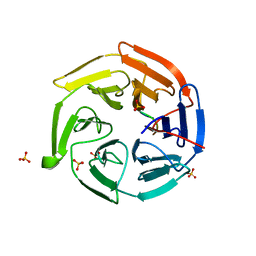 | |
2E3K
 
 | |
7ENZ
 
 | | Crystal structure of Phenanthredinone moiety in complex with the second bromodomain of BRD2 (BRD2-BD2). | | Descriptor: | Bromodomain-containing protein 2, TRIETHYLENE GLYCOL, phenanthridin-6(5H)-one | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7XX3
 
 | | Crystal structure of human Superoxide Dismutase (SOD1) in complex with a fungal metabolite molecule, Phialomustin B (PB) | | Descriptor: | (2~{E},4~{E},6~{S})-4,6-dimethyldeca-2,4-dienoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Padmanabhan, B, Unni, S. | | Deposit date: | 2022-05-28 | | Release date: | 2023-12-13 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the modulation Of SOD1 aggregation By a fungal metabolite Phialomustin-B: Therapeutic potential in ALS.
Plos One, 19, 2024
|
|
5XHE
 
 | | Crystal structure analysis of the second bromodomain of BRD2 covalently linked to b-mercaptoethanol | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, TRIETHYLENE GLYCOL | | Authors: | Padmanabhan, B, Mathur, S, Tripathi, S.K, Deshmukh, P. | | Deposit date: | 2017-04-20 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the crystal structure of BRD2-BD2 - phenanthridinone complex and theoretical studies on phenanthridinone analogs.
J. Biomol. Struct. Dyn., 36, 2018
|
|
5XHK
 
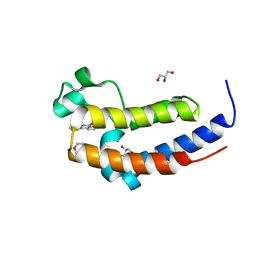 | | Crystal structure of the BRD2-BD2 in complex with phenanthridinone | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, METHOXYETHANE, ... | | Authors: | Padmanabhan, B, Mathur, S, Tripathi, S, Deshmukh, P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Insights into the crystal structure of BRD2-BD2 - phenanthridinone complex and theoretical studies on phenanthridinone analogs.
J. Biomol. Struct. Dyn., 36, 2018
|
|
