5BT3
 
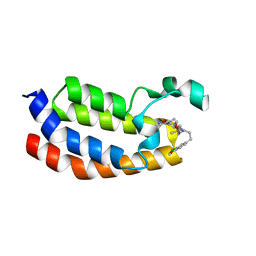 | | Crystal structure of EP300 bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Histone acetyltransferase p300, ISOPROPYL ALCOHOL | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of EP300 bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5CC9
 
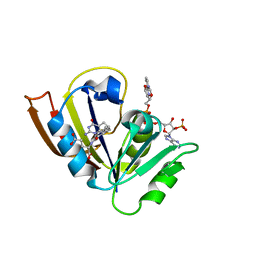 | |
5CCC
 
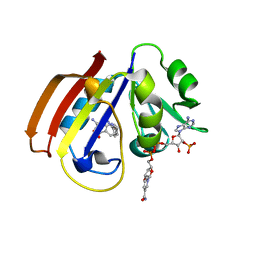 | |
5E9V
 
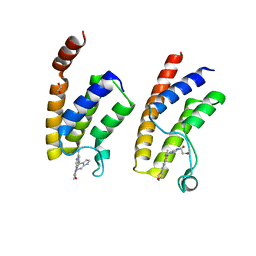 | | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(imidazo[1,2-a]pyridin-5-yl)-7-(morpholin-4-yl)indolizin-3-yl]ethanone, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Hay, D.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Schofield, C.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand
To Be Published
|
|
1PRT
 
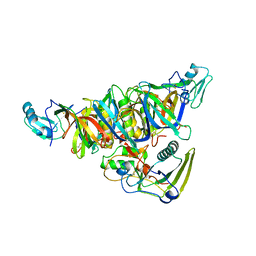 | | THE CRYSTAL STRUCTURE OF PERTUSSIS TOXIN | | Descriptor: | PERTUSSIS TOXIN (SUBUNIT S1), PERTUSSIS TOXIN (SUBUNIT S2), PERTUSSIS TOXIN (SUBUNIT S3), ... | | Authors: | Stein, P.E, Read, R.J. | | Deposit date: | 1993-11-22 | | Release date: | 1995-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of pertussis toxin.
Structure, 2, 1994
|
|
1POU
 
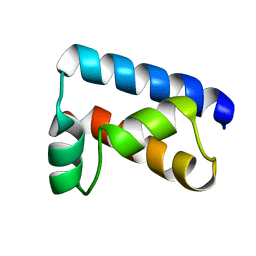 | | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | | Descriptor: | OCT-1 | | Authors: | Assa-Munt, N, Mortishire-Smith, R.J, Aurora, R, Herr, W, Wright, P.E. | | Deposit date: | 1993-06-14 | | Release date: | 1994-10-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Cell(Cambridge,Mass.), 73, 1993
|
|
1QPD
 
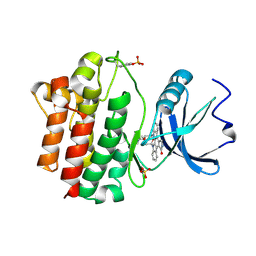 | | STRUCTURAL ANALYSIS OF THE LYMPHOCYTE-SPECIFIC KINASE LCK IN COMPLEX WITH NON-SELECTIVE AND SRC FAMILY SELECTIVE KINASE INHIBITORS | | Descriptor: | LCK KINASE, STAUROSPORINE, SULFATE ION | | Authors: | Zhu, X, Kim, J.L, Rose, P.E, Stover, D.R, Toledo, L.M, Zhao, H, Morgenstern, K.A. | | Deposit date: | 1999-05-24 | | Release date: | 2000-05-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors.
Structure Fold.Des., 7, 1999
|
|
1QPJ
 
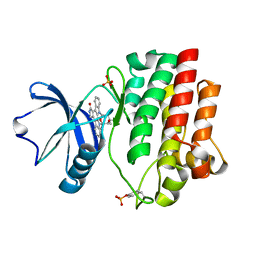 | | CRYSTAL STRUCTURE OF THE LYMPHOCYTE-SPECIFIC KINASE LCK IN COMPLEX WITH STAUROSPORINE. | | Descriptor: | LCK TYROSINE KINASE, STAUROSPORINE, SULFATE ION | | Authors: | Zhu, X, Kim, J.L, Rose, P.E, Stover, D.R, Toledo, L.M. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors.
Structure Fold.Des., 7, 1999
|
|
1QPC
 
 | | STRUCTURAL ANALYSIS OF THE LYMPHOCYTE-SPECIFIC KINASE LCK IN COMPLEX WITH NON-SELECTIVE AND SRC FAMILY SELECTIVE KINASE INHIBITORS | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Zhu, X, Kim, J.L, Rose, P.E, Stover, D.R, Toledo, L.M, Zhao, H, Morgenstern, K.A. | | Deposit date: | 1999-05-21 | | Release date: | 2000-05-24 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors.
Structure Fold.Des., 7, 1999
|
|
1R8U
 
 | | NMR structure of CBP TAZ1/CITED2 complex | | Descriptor: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | Authors: | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-03-23 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
1R1L
 
 | | Structure of dimeric antithrombin complexed with a P14-P9 reactive loop peptide and an exogenous tripeptide (formyl-norleucine-LF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin P14-P9 peptide, Antithrombin-III, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Stein, P.E, Carrell, R.W. | | Deposit date: | 2003-09-24 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Serpins and the design of peptides to block intermolecular beta-linkages
To be Published
|
|
1R5E
 
 | | Solution structure of the folded core of Pseudomonas syringae effector protein, AvrPto | | Descriptor: | avirulence protein | | Authors: | Wulf, J, Pascuzzi, P.E, Martin, G.B, Nicholson, L.K. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-19 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis.
STRUCTURE, 12, 2004
|
|
1RGX
 
 | | Crystal Structure of resisitin | | Descriptor: | Resistin | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
1RH7
 
 | | Crystal Structure of Resistin-like beta | | Descriptor: | HEXAETHYLENE GLYCOL, PLATINUM (II) ION, Resistin-like beta | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
1RFX
 
 | | Crystal Structure of resisitin | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-10 | | Release date: | 2004-06-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
1RGO
 
 | | Structural Basis for Recognition of the mRNA Class II AU-Rich Element by the Tandem Zinc Finger Domain of TIS11d | | Descriptor: | Butyrate response factor 2, RNA (5'-R(*UP*UP*AP*UP*UP*UP*AP*UP*U)-3'), ZINC ION | | Authors: | Hudson, B.P, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d.
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1RXR
 
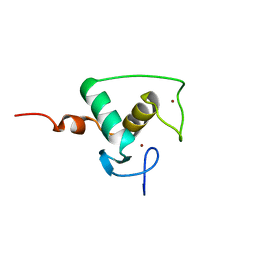 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE RETINOID X RECEPTOR DNA BINDING DOMAIN, NMR, 20 STRUCTURE | | Descriptor: | RETINOIC ACID RECEPTOR-ALPHA, ZINC ION | | Authors: | Holmbeck, S.M.A, Foster, M.P, Casimiro, D.R, Sem, D.S, Dyson, H.J, Wright, P.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the retinoid X receptor DNA-binding domain.
J.Mol.Biol., 281, 1998
|
|
1SFF
 
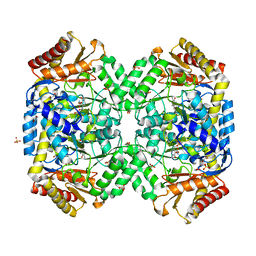 | | Structure of gamma-aminobutyrate aminotransferase complex with aminooxyacetate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
1SB0
 
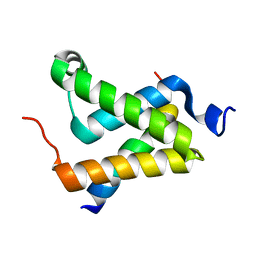 | | Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb | | Descriptor: | protein CBP, protein c-Myb | | Authors: | Zor, T, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the KIX Domain of CBP Bound to the Transactivation
Domain of c-Myb
J.Mol.Biol., 337, 2004
|
|
1SF2
 
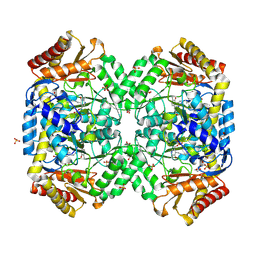 | | Structure of E. coli gamma-aminobutyrate aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
1S88
 
 | | NMR structure of a DNA duplex with two INA nucleotides inserted opposite each other, dCTCAACXCAAGCT:dAGCTTGXGTTGAG | | Descriptor: | 5'-D(*AP*GP*CP*TP*TP*GP*(2DM)P*GP*TP*TP*GP*AP*G)-3', 5'-D(*CP*TP*CP*AP*AP*CP*(2DM)P*CP*AP*AP*GP*CP*T)-3' | | Authors: | Nielsen, C.B, Petersen, M, Pedersen, E.B, Hansen, P.E, Christensen, U.B. | | Deposit date: | 2004-01-31 | | Release date: | 2004-05-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a modified DNA oligonucleotide containing a new intercalating nucleic acid.
Bioconjug.Chem., 15, 2004
|
|
1SZK
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: E211S | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1SZS
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: I50Q | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1T56
 
 | | Crystal structure of TetR family repressor M. tuberculosis EthR | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, EthR repressor, GLYCEROL | | Authors: | Dover, L.G, Corsino, P.E, Daniels, I.R, Cocklin, S.L, Tatituri, V, Besra, G.S, Futterer, K. | | Deposit date: | 2004-05-03 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the TetR/CamR Family Repressor Mycobacterium tuberculosis EthR Implicated in Ethionamide Resistance.
J.Mol.Biol., 340, 2004
|
|
1SZU
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
