1VA2
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA3
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA1
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1ISE
 
 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | Descriptor: | Ribosome Recycling Factor | | Authors: | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
3W39
 
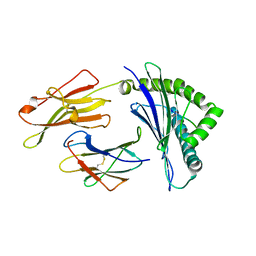 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
2D0J
 
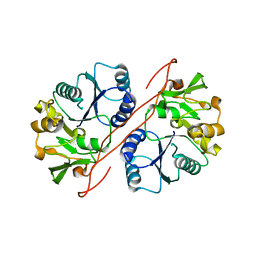 | | Crystal Structure of Human GlcAT-S Apo Form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 | | Authors: | Shiba, T, Kakuda, S, Ishiguro, M, Oka, S, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-08-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GlcAT-S, a human glucuronyltransferase, involved in the biosynthesis of the HNK-1 carbohydrate epitope
To be published
|
|
1V84
 
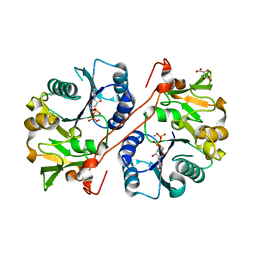 | | Crystal structure of human GlcAT-P in complex with N-acetyllactosamine, Udp, and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V83
 
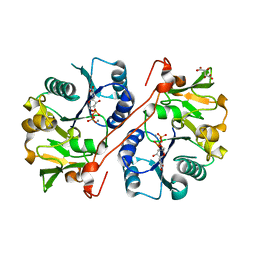 | | Crystal structure of human GlcAT-P in complex with Udp and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V82
 
 | | Crystal structure of human GlcAT-P apo form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
4MJI
 
 | | T cell response to a HIV reverse transcriptase epitope presented by the protective allele HLA-B*51:01 | | Descriptor: | Beta-2-microglobulin, HIV Reverse Transcriptase peptide Marker, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Motozono, C, Takiguchi, M. | | Deposit date: | 2013-09-03 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Molecular basis of a dominant T cell response to an HIV reverse transcriptase 8-mer epitope presented by the protective allele HLA-B*51:01
J.Immunol., 192, 2014
|
|
4DG1
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
5HGB
 
 | | HLA*A2402 complexed with HIV nef138 8mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
5HGH
 
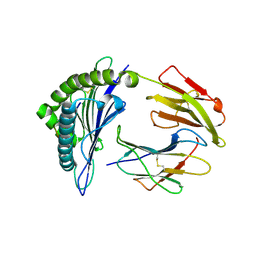 | | HLA*A2402 complexed with HIV nef138 10mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
5HGD
 
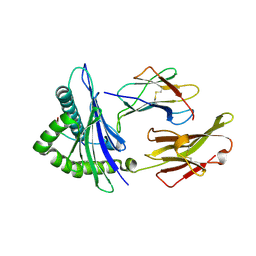 | | HLA*A2402 complexed with HIV nef138 Y2F mutant 10mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
5HGA
 
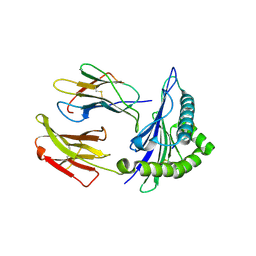 | | HLA*A2402 complex with HIV nef138 Y2F-8mer mutant epitope | | Descriptor: | 8-mer from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
