7ZEN
 
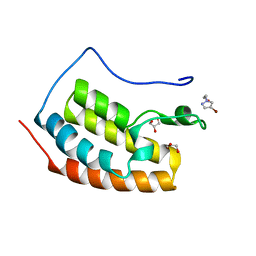 | | BRD4 in complex with FragLite23 | | Descriptor: | 2-(4-bromo-1H-pyrazol-1-yl)ethan-1-ol, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFN
 
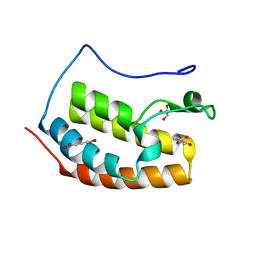 | | BRD4 in complex with FragLite24 | | Descriptor: | 4-bromanyl-2-oxidanyl-benzoic acid, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFT
 
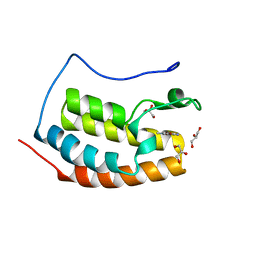 | | BRD4 in complex with FragLite33 | | Descriptor: | 3-azanyl-5-bromanyl-1-methyl-pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZA7
 
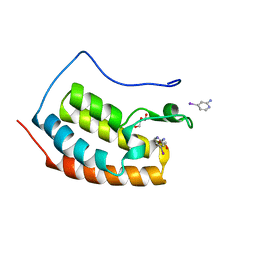 | | BRD4 in complex with FragLite5 | | Descriptor: | 4-iodanylpyridin-2-amine, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZAA
 
 | | BRD4 in complex with FragLite11 | | Descriptor: | GLYCEROL, Isoform C of Bromodomain-containing protein 4, ~{N}-(4-bromophenyl)ethanamide | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZAT
 
 | | BRD4 in complex with FragLite20 | | Descriptor: | 4-bromanyl-2-(methoxymethyl)pyridine, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZAE
 
 | | BRD4 in complex with FragLite15 | | Descriptor: | 4-bromanyl-2-methoxy-pyridine, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFY
 
 | | BRD4 in complex with PepLite-Gly | | Descriptor: | 2-acetamido-N-(3-bromanylprop-2-ynyl)ethanamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZG2
 
 | | BRD4 in complex with Acetyl-Lys | | Descriptor: | (2S)-2,6-diacetamido-N-methyl-hexanamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZA8
 
 | | BRD4 in complex with FragLite6 | | Descriptor: | 4-bromanyl-1~{H}-pyridin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7Z9W
 
 | | BRD4 in complex with FragLite1 | | Descriptor: | 4-bromo-1H-pyrazole, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.988 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZE7
 
 | | BRD4 in complex with FragLite21 | | Descriptor: | 5-bromo-2-hydroxybenzonitrile, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7Z9Y
 
 | | BRD4 in complex with FragLite2 | | Descriptor: | 4-IODOPYRAZOLE, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
1W5R
 
 | | X-ray crystallographic structure of a C70Q Mycobacterium smegmatis N- arylamine Acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Holton, S.J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E.M, Sim, E. | | Deposit date: | 2004-08-09 | | Release date: | 2005-05-11 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Investigation of the Catalytic Triad of Arylamine N-Acetyltransferases: Essential Residues Required for Acetyl Transfer to Arylamines.
Biochem.J., 390, 2005
|
|
1W6F
 
 | | Arylamine N-acetyltransferase from Mycobacterium smegmatis with the anti-tubercular drug isoniazid bound in the active site. | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Holton, S, Fullham, E, Sim, E, Noble, M.E.M. | | Deposit date: | 2004-08-17 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Anti-Tubercular Drug Isoniazid to the Arylamine N-Acetyltransferase Protein from Mycobacterium Smegmatis
Protein Sci., 14, 2005
|
|
5NZQ
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-(1,3-oxazol-5-yl)aniline. | | Descriptor: | 3-(1,3-oxazol-5-yl)aniline, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5NZP
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-Hydroxybenzisoxazole | | Descriptor: | 1,2-benzoxazol-3-ol, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5OFV
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-fluoro-2-methylbenzoic acid | | Descriptor: | 5-fluoranyl-2-methyl-benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5NZO
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 1-methyl-3-phenyl-1H-pyrazol-5-amine | | Descriptor: | 2-methyl-5-phenyl-pyrazol-3-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5OFW
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-Chloro-4-fluorobenzamide | | Descriptor: | 3-chloranyl-4-fluoranyl-benzamide, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5OFM
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole | | Descriptor: | 1-methylindol-5-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole
To be published
|
|
4HG7
 
 | | Crystal Structure of an MDM2/Nutlin-3a complex | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Noble, M.E.M, Anil, B, Riedinger, C, Endicott, J.A. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1TAH
 
 | | THE CRYSTAL STRUCTURE OF TRIACYLGLYCEROL LIPASE FROM PSEUDOMONAS GLUMAE REVEALS A PARTIALLY REDUNDANT CATALYTIC ASPARTATE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Noble, M.E.M, Cleasby, A, Johnson, L.N, Egmond, M, Frenken, L.G.J. | | Deposit date: | 1993-12-21 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate.
FEBS Lett., 331, 1993
|
|
1TRD
 
 | |
1TPD
 
 | |
