6A6H
 
 | |
8TN8
 
 | |
3U1H
 
 | |
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
5JAE
 
 | | LeuT in the outward-oriented, Na+-free return state, P21 form at pH 6.5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
4UQI
 
 | | AP2 controls clathrin polymerization with a membrane-activated switch | | Descriptor: | AP-2 COMPLEX SUBUNIT ALPHA-2, AP-2 COMPLEX SUBUNIT BETA, AP-2 COMPLEX SUBUNIT MU, ... | | Authors: | Kelly, B.T, Graham, S.C, Liska, N, Dannhauser, P.N, Hoening, S, Ungewickell, E.J, Owen, D.J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Clathrin Adaptors. Ap2 Controls Clathrin Polymerization with a Membrane-Activated Switch.
Science, 345, 2014
|
|
1AIH
 
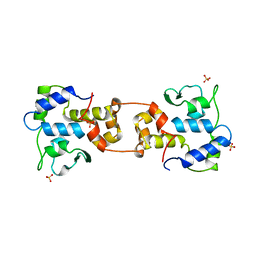 | | CATALYTIC DOMAIN OF BACTERIOPHAGE HP1 INTEGRASE | | Descriptor: | HP1 INTEGRASE, MAGNESIUM ION, SULFATE ION | | Authors: | Hickman, A.B, Waninger, S, Scocca, J.J, Dyda, F. | | Deposit date: | 1997-04-17 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Cell(Cambridge,Mass.), 89, 1997
|
|
5JAG
 
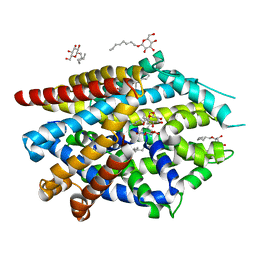 | | LeuT T354H mutant in the outward-oriented, Na+-free Return State | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
5JAF
 
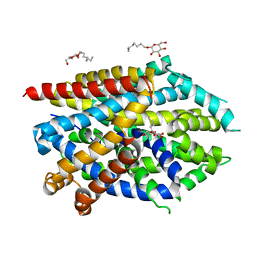 | | LeuT Na+-free Return State, C2 form at pH 5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.021 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
3C0T
 
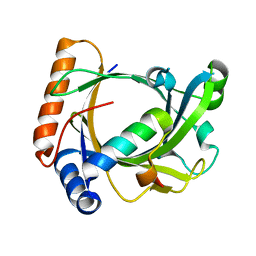 | | Structure of the Schizosaccharomyces pombe Mediator subcomplex Med8C/18 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 18, Mediator of RNA polymerase II transcription subunit 8 | | Authors: | Lariviere, L, Seizl, M, van Wageningen, S, Roether, S, van de Pasch, L, Feldmann, H, Straesser, K, Hahn, S, Holstege, C.P, Cramer, P. | | Deposit date: | 2008-01-21 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-system correlation identifies a gene regulatory Mediator submodule
Genes Dev., 22, 2008
|
|
4CCA
 
 | | Structure of human Munc18-2 | | Descriptor: | CHLORIDE ION, SYNTAXIN-BINDING PROTEIN 2 | | Authors: | Hackmann, Y, Graham, S.C, Ehl, S, Hoening, S, Lehmberg, K, Arico, M, Owen, D.J, Griffiths, G.G. | | Deposit date: | 2013-10-21 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Syntaxin Binding Mechanism and Disease-Causing Mutations in Munc18-2
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ATJ
 
 | | DISTAL HEME POCKET MUTANT (H42E) OF RECOMBINANT HORSERADISH PEROXIDASE IN COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PROTEIN (PEROXIDASE C1A), ... | | Authors: | Meno, K, Jennings, S, Smith, A.T, Henriksen, A, Gajhede, M. | | Deposit date: | 1999-04-19 | | Release date: | 2002-10-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3ZYK
 
 | | Structure of CALM (PICALM) ANTH domain | | Descriptor: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYM
 
 | | Structure of CALM (PICALM) in complex with VAMP8 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN, ... | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYL
 
 | | Structure of a truncated CALM (PICALM) ANTH domain | | Descriptor: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
6ZBV
 
 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor and sybody Sb_GlyT1#7 | | Descriptor: | Sodium- and chloride-dependent glycine transporter 1,Sodium- and chloride-dependent glycine transporter 1, Sybody Sb_GlyT1#7, [5-fluoranyl-6-(oxan-4-yloxy)-1,3-dihydroisoindol-2-yl]-[5-methylsulfonyl-2-[2,2,3,3,3-pentakis(fluoranyl)propoxy]phenyl]methanone | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
6ZPL
 
 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
7A8P
 
 | | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | | Descriptor: | (3~{R})-1-[(2~{R})-2-[4-(2-chloranyl-4-fluoranyl-phenyl)-2-oxidanylidene-chromen-7-yl]oxypropanoyl]piperidine-3-carboxylic acid, DNA-directed RNA polymerase, mitochondrial | | Authors: | Hillen, H.S, Bonekamp, N, Peter, B, Felser, A, Bergbrede, T, Choidas, A, Horn, M, Unger, A, di Lucrezia, R, Atanassov, I, Li, X, Koch, U, Menninger, S, Boros, J, Habenberger, P, Giavalisco, P, Cramer, P, Denzel, M, Nussbaumer, P, Klebl, B, Falkenberg, M, Gustafsson, C.M, Larsson, N.G. | | Deposit date: | 2020-08-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Small-molecule inhibitors of human mitochondrial DNA transcription.
Nature, 588, 2020
|
|
6XDG
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
3EO1
 
 | |
3EO0
 
 | |
8OZ6
 
 | | cryoEM structure of SPARTA complex ligand-free | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZF
 
 | | cryoEM structure of SPARTA complex Tetramer Post-NAD cleavage-2 | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OV5
 
 | |
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
