5K4H
 
 | | Wolinella succinogenes L-asparaginase S121 + L-Glutamic acid | | Descriptor: | GLUTAMIC ACID, L-asparaginase | | Authors: | Nguyen, H.A, Lave, A. | | Deposit date: | 2016-05-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The differential ability of asparagine and glutamine in promoting the closed/active enzyme conformation rationalizes the Wolinella succinogenes L-asparaginase substrate specificity.
Sci Rep, 7, 2017
|
|
5K4G
 
 | |
5HW0
 
 | |
5I4B
 
 | |
5I3Z
 
 | |
5I48
 
 | |
5K3O
 
 | | Wolinella succinogenes L-asparaginase P121 and L-Aspartic acid | | Descriptor: | ASPARTIC ACID, L-asparaginase | | Authors: | Nguyen, H.A, Lave, A. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | The differential ability of asparagine and glutamine in promoting the closed/active enzyme conformation rationalizes the Wolinella succinogenes L-asparaginase substrate specificity.
Sci Rep, 7, 2017
|
|
5K45
 
 | | Wolinella succinogenes L-asparaginase P121 + L-Glutamic acid | | Descriptor: | GLUTAMIC ACID, L-asparaginase | | Authors: | Nguyen, H.A, Lave, A. | | Deposit date: | 2016-05-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The differential ability of asparagine and glutamine in promoting the closed/active enzyme conformation rationalizes the Wolinella succinogenes L-asparaginase substrate specificity.
Sci Rep, 7, 2017
|
|
5F52
 
 | | Erwinia chrysanthemi L-asparaginase + Aspartic acid | | Descriptor: | ASPARTIC ACID, DI(HYDROXYETHYL)ETHER, L-asparaginase | | Authors: | Nguyen, H.A, Lavie, A. | | Deposit date: | 2015-12-04 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Insight into Substrate Selectivity of Erwinia chrysanthemi l-Asparaginase.
Biochemistry, 55, 2016
|
|
6OF6
 
 | | Crystal structure of tRNA^ Ala(GGC) bound to cognate 70S A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OPE
 
 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to near-cognate 70S A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-04-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FOM
 
 | | Crystal structure of tRNA^Lys(SUU) bound to UAA codon in the ribosomal P site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2023-01-02 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
8FON
 
 | | Crystal structure of tRNA^Lys(SUU) bound to AUA codon in the ribosomal P site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2023-01-02 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
6ORD
 
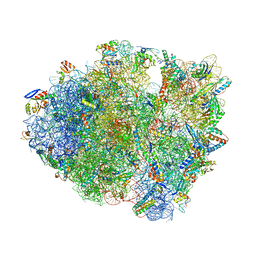 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to cognate 70S A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OJ2
 
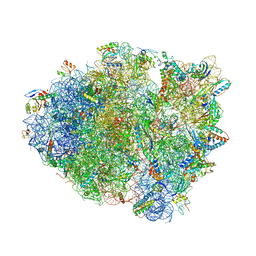 | | Crystal structure of tRNA^ Ala(GGC) bound to the near-cognate 70S A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-04-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FR3
 
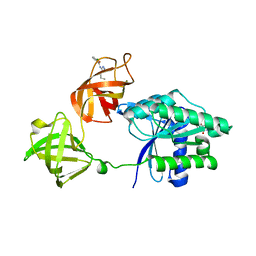 | | E. coli EF-Tu in complex with KKL-55 | | Descriptor: | 3-chloro-N-(1-propyl-1H-tetrazol-5-yl)benzamide, Elongation factor Tu, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nguyen, H.A, Kuzmishin Nagy, A.B, Dunham, C.M. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Antibiotic that inhibits trans -translation blocks binding of EF-Tu to tmRNA but not to tRNA.
Mbio, 14, 2023
|
|
7U6M
 
 | |
5NP9
 
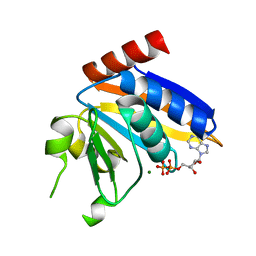 | |
5MVR
 
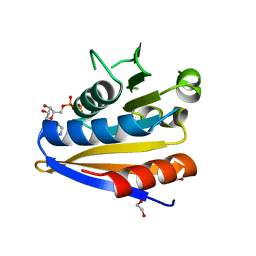 | | Crystal structure of Bacillus subtilus YdiB | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jault, J.-M, Aghajari, N. | | Deposit date: | 2017-01-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Expanding the Kinome World: A New Protein Kinase Family Widely Conserved in Bacteria.
J. Mol. Biol., 429, 2017
|
|
6NDK
 
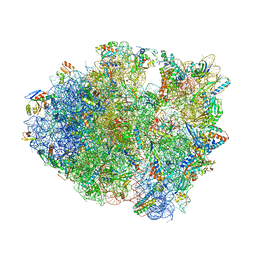 | | Structure of ASLSufA6 A37.5 bound to the 70S A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.T, Hoffer, E.D, Dunham, C.M. | | Deposit date: | 2018-12-13 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Importance of a tRNA anticodon loop modification and a conserved, noncanonical anticodon stem pairing intRNACGGProfor decoding
J. Biol. Chem., 294, 2019
|
|
7S0S
 
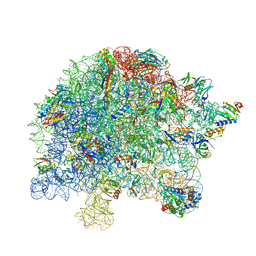 | | M. tuberculosis ribosomal RNA methyltransferase TlyA bound to M. smegmatis 50S ribosomal subunit | | Descriptor: | 16S/23S rRNA (Cytidine-2'-O)-methyltransferase TlyA, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Laughlin, Z.T, Dunham, C.M, Conn, G.L. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | 50S subunit recognition and modification by the Mycobacterium tuberculosis ribosomal RNA methyltransferase TlyA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4Q1B
 
 | |
4Q1D
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 9 {2-{[(1R)-1-{2-[3-(2-fluoroethoxy)-4-methoxyphenyl]-5-propyl-1,3-thiazol-4-yl}ethyl]sulfanyl}pyrimidine-4,6-diamine} | | Descriptor: | (R)-2-((1-(2-(3-(2-fluoroethoxy)-4-methoxyphenyl)-5-propylthiazol-4-yl)ethyl)thio)pyrimidine-4,6-diamine, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
4Q19
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 5 {5-(4-{[(4,6-DIAMINOPYRIMIDIN-2-YL)SULFANYL]METHYL}-5-PROPYL-1,3-THIAZOL-2-YL)-2-METHOXYPHENOL} | | Descriptor: | 5-(4-(((4,6-diaminopyrimidin-2-yl)thio)methyl)-5-propylthiazol-2-yl)-2-methoxyphenol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
4Q18
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 4 [1-[5-(4-{[(2,6-diaminopyrimidin-4-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]-2-methylpropan-2-ol] | | Descriptor: | 1-(5-(4-(((2,6-diaminopyrimidin-4-yl)thio)methyl)-5-propylthiazol-2-yl)-2-methoxyphenoxy)-2-methylpropan-2-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
