3LQ2
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with low TDP concentration | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
3LQ4
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with high TDP concentration | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
3LPL
 
 | | E. coli pyruvate dehydrogenase complex E1 component E571A mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
6U3J
 
 | | Structure of the 2-oxoadipate dehydrogenase DHTKD1 | | Descriptor: | 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, MAGNESIUM ION, ... | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2019-08-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Inhibition and Crystal Structure of the Human DHTKD1-Thiamin Diphosphate Complex.
Acs Chem.Biol., 15, 2020
|
|
6CFO
 
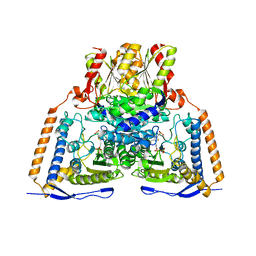 | | HUMAN PYRUVATE DEHYDROGENASE E1 COMPONENT COMPLEX WITH COVALENT TDP ADDUCT ACETYL PHOSPHINATE | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(1S)-1-hydroxy-1-[(R)-hydroxy(oxo)-lambda~5~-phosphanyl]ethyl}-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, MAGNESIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | ARJUNAN, P, WHITLEY, M.J, FUREY, W. | | Deposit date: | 2018-02-15 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyruvate dehydrogenase complex deficiency is linked to regulatory loop disorder in the alpha V138M variant of human pyruvate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
4JDR
 
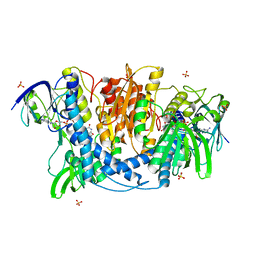 | | Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase from escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chandrasekhar, K, Arjunan, P, Furey, W. | | Deposit date: | 2013-02-25 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight to the Interaction of the Dihydrolipoamide Acetyltransferase (E2) Core with the Peripheral Components in the Escherichia coli Pyruvate Dehydrogenase Complex via Multifaceted Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
3F6B
 
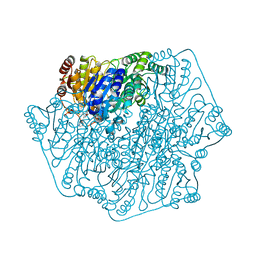 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor PAA | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
6CER
 
 | | Human pyruvate dehydrogenase complex E1 component V138M mutation | | Descriptor: | MAGNESIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Whitley, M.J, Arjunan, P, Furey, W. | | Deposit date: | 2018-02-12 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Pyruvate dehydrogenase complex deficiency is linked to regulatory loop disorder in the alpha V138M variant of human pyruvate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
3F6E
 
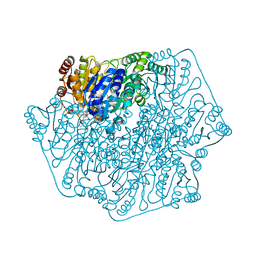 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor 3-PKB | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
4N72
 
 | |
4QOY
 
 | |
6H05
 
 | | Cryo-electron microscopic structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate (2-oxoglutarate) dehydrogenase complex [residues 218-453] | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Nagy, B, Zambo, Z, Hubert, A, Polak, M, Nemeria, N.S, Novacek, J, Jordan, F, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-07-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: Implications for the overall hKGDHc structure.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
