1WSV
 
 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-11 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1X1N
 
 | | Structure determination and refinement at 1.8 A resolution of Disproportionating Enzyme from Potato | | Descriptor: | 4-alpha-glucanotransferase, CALCIUM ION, GLYCEROL | | Authors: | Imamura, K, Matsuura, T, Takaha, T, Fujii, K, Nakagawa, A, Kusunoki, M, Nitta, Y. | | Deposit date: | 2005-04-08 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination and refinement at 1.8 A resolution of Disproportionating Enzyme from Potato
to be published
|
|
1WSR
 
 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1X2H
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1X2G
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli | | Descriptor: | Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1Y43
 
 | | crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | Descriptor: | Aspergillopepsin II heavy chain, Aspergillopepsin II light chain, SULFATE ION | | Authors: | Sasaki, H, Nakagawa, A, Iwata, S, Muramatsu, T, Suganuma, M, Sawano, Y, Kojima, M, Kubota, K, Takahashi, K. | | Deposit date: | 2004-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The three-dimensional structure of aspergilloglutamic peptidase from Aspergillus niger
Proc.Jpn.Acad.,Ser.B, 80, 2004
|
|
6BWU
 
 | | Crystal structure of carboxyhemoglobin in complex with beta Cys93 modifying agent, TD3 | | Descriptor: | 1H-1,2,3-triazole-5-thiol, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Musayev, F.N, Safo, R.M, Safo, M.K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Mol. Pharm., 15, 2018
|
|
6BWP
 
 | | Crystal structure of Deoxy Hemoglobin in complex with beta Cys93 modifying agent, TD3 | | Descriptor: | 1H-1,2,3-triazole-5-thiol, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Musayev, F.N, Safo, R.M, Safo, M.K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Mol. Pharm., 15, 2018
|
|
4WIZ
 
 | | Crystal structure of Grouper nervous necrosis virus-like particle at 3.6A | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
487D
 
 | |
4QN0
 
 | | Crystal structure of the CPS-6 mutant Q130K | | Descriptor: | Endonuclease G, mitochondrial, MAGNESIUM ION | | Authors: | Lin, J.L.J, Yuan, H.S. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Oxidative Stress Impairs Cell Death by Repressing the Nuclease Activity of Mitochondrial Endonuclease G
Cell Rep, 16, 2016
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZW
 
 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
8JU3
 
 | | Mu phage tail fiber | | Descriptor: | Tail fiber protein S,Tail fiber protein S' | | Authors: | Yamashita, E, Takeda, S. | | Deposit date: | 2023-06-24 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of three-dimensional structure of bacteriophase Mu tail fiber
To Be Published
|
|
5B82
 
 | |
4QPD
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 10-formyltetrahydrofolate dehydrogenase, DI(HYDROXYETHYL)ETHER | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R8V
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RFU
 
 | | Crystal structure of truncated P-domain from Grouper nervous necrosis virus capsid protein at 1.2A | | Descriptor: | CALCIUM ION, Coat protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
4RFT
 
 | | T=1 subviral particle of Grouper nervous necrosis virus capsid protein deletion mutant (delta 1-34 & 218-338) | | Descriptor: | Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
4TT8
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TTS
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TS4
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) from zebrafish | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4L95
 
 | | Crystal structure of gamma glutamyl hydrolase (H218N) from zebrafish | | Descriptor: | GLYCEROL, Gamma-glutamyl hydrolase | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L8W
 
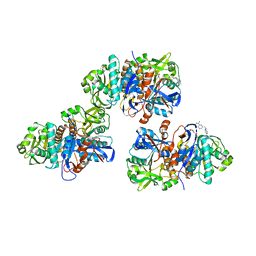 | | Crystal structure of gamma glutamyl hydrolase (H218N) from zebrafish complex with MTX polyglutamate | | Descriptor: | D-GLUTAMIC ACID, Gamma-glutamyl hydrolase, METHOTREXATE | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
