5OGA
 
 | | Structure of minimal i-motif domain | | Descriptor: | DNA (5'-D(*TP*(DCP)P*GP*TP*TP*CP*(DCP)P*GP*TP*TP*TP*TP*TP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Mir, B, Serrano, I, Buitrago, D, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2017-07-12 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Prevalent Sequences in the Human Genome Can Form Mini i-Motif Structures at Physiological pH.
J. Am. Chem. Soc., 139, 2017
|
|
5NIP
 
 | |
8OFC
 
 | | Structure of an i-motif domain with the cytosine analog 1,3-diaza-2-oxophenoxacione (tC) at neutral pH | | Descriptor: | DNA (5'-D(*CP*(YCO)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*(DNR)P*GP*T)-3') | | Authors: | Mir, B, Serrano-Chacon, I, Terrazas, M, Gandioso, A, Garavis, M, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2023-03-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Site-specific incorporation of a fluorescent nucleobase analog enhances i-motif stability and allows monitoring of i-motif folding inside cells.
Nucleic Acids Res., 52, 2024
|
|
3AI8
 
 | | Cathepsin B in complex with the nitroxoline | | Descriptor: | 5-nitroquinolin-8-ol, Cathepsin B | | Authors: | Renko, M, Mirkovic, B, Gobec, S, Kos, J, Turk, D. | | Deposit date: | 2010-05-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Novel mechanism of cathepsin B inhibition by antibiotic nitroxoline and related compounds
Chemmedchem, 6, 2011
|
|
7O5E
 
 | |
8BQY
 
 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BV6
 
 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
7SBG
 
 | |
7SBD
 
 | |
7SD2
 
 | |
8G45
 
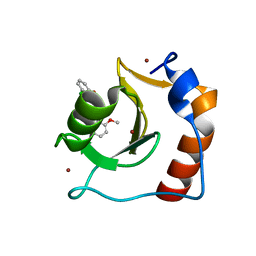 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with SGC-UBD253 chemical probe | | Descriptor: | 3-[8-chloro-3-(2-{[(2-methoxyphenyl)methyl]amino}-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl]propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G43
 
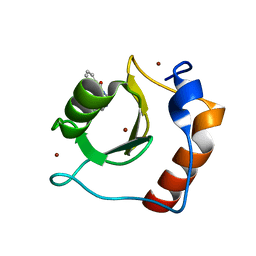 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G44
 
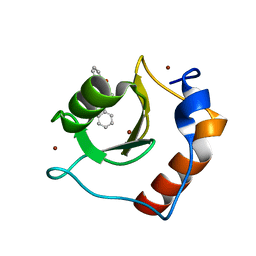 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(benzylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(benzylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8UWU
 
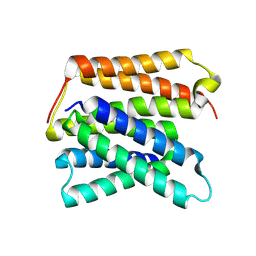 | | EmrE structure in the proton-bound state (WT/L51I heterodimer) | | Descriptor: | SMR family multidrug efflux protein EmrE | | Authors: | Li, J, Sae Her, A, Besch, A, Ramirez, B, Crames, M, Banigan, J.R, Mueller, C, Marsiglia, W.M, Zhang, Y, Traaseth, N.J. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Nat Commun, 15, 2024
|
|
8UOZ
 
 | | EmrE structure in the TPP-bound state (WT/E14Q heterodimer) | | Descriptor: | SMR family multidrug efflux protein EmrE, TETRAPHENYLPHOSPHONIUM | | Authors: | Li, J, Sae Her, A, Besch, A, Ramirez, B, Crames, M, Banigan, J.R, Mueller, C, Marsiglia, W.M, Zhang, Y, Traaseth, N.J. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Nat Commun, 15, 2024
|
|
6UL3
 
 | |
6UKC
 
 | |
1VJI
 
 | | Gene Product of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase (OPR1), FLAVIN MONONUCLEOTIDE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1.
Proteins, 61, 2005
|
|
3S5A
 
 | | ABH2 cross-linked to undamaged dsDNA-2 with cofactors | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*CP*AP*CP*TP*GP*TP*CP*G)-3', 5'-D(*TP*CP*GP*AP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Ramirez, B, Zhang, W, Jia, G, Zhang, L, Li, C.Q, Dinner, A.R, Yang, C.-G, He, C. | | Deposit date: | 2011-05-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3S57
 
 | | ABH2 cross-linked with undamaged dsDNA-1 containing cofactors | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*AP*TP*CP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*TP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Ramirez, B, Zhang, W, Jia, G, Zhang, L, Li, C.Q, Dinner, A.R, Yang, C.-G, He, C. | | Deposit date: | 2011-05-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
2M9U
 
 | | Solution NMR structure of the C-terminal domain (CTD) of Moloney murine leukemia virus integrase, Northeast Structural Genomics Target OR41A | | Descriptor: | Integrase p46 | | Authors: | Aiyer, S, Rossi, P, Schneider, W.M, Chander, A, Roth, M.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction.
Nucleic Acids Res., 42, 2014
|
|
