1GVN
 
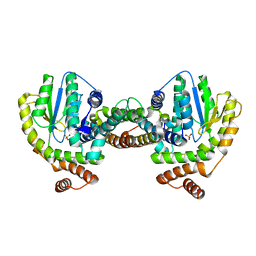 | | Crystal Structure of the Plasmid Maintenance System epsilon/zeta: Meachnism of toxin inactivation and toxin function | | Descriptor: | EPSILON, SULFATE ION, ZETA | | Authors: | Meinhart, A, Alonso, J.C, Straeter, N, Saenger, W. | | Deposit date: | 2002-02-19 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Plasmid Maintenance System Epsilon /Zeta : Functional Mechanism of Toxin Zeta and Inactivation by Epsilon 2 Zeta 2 Complex Formation
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Q1H
 
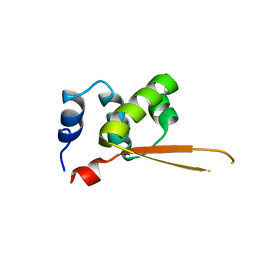 | |
1SZA
 
 | |
1SZ9
 
 | |
6ZK7
 
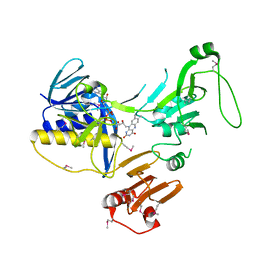 | | Crystal Structure of human PYROXD1/FAD complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase domain-containing protein 1 | | Authors: | Meinhart, A, Asanovic, I, Martinez, J, Clausen, T. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The oxidoreductase PYROXD1 uses NAD(P) + as an antioxidant to sustain tRNA ligase activity in pre-tRNA splicing and unfolded protein response.
Mol.Cell, 81, 2021
|
|
6QDJ
 
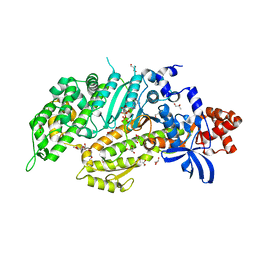 | | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin | | Descriptor: | 1,4-BUTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Meinhart, A, Clausen, T, Arnese, R. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin.
Nat Commun, 10, 2019
|
|
6QDL
 
 | |
6QDM
 
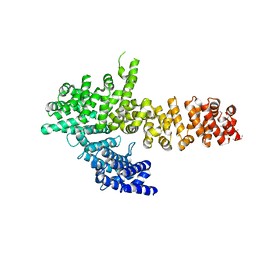 | |
6QDK
 
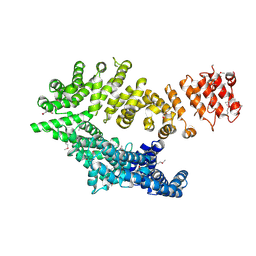 | |
7AA4
 
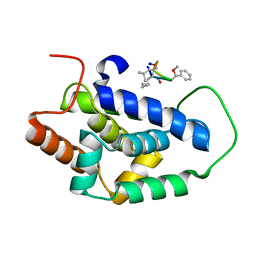 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
8B9O
 
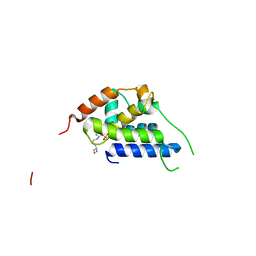 | |
8B9U
 
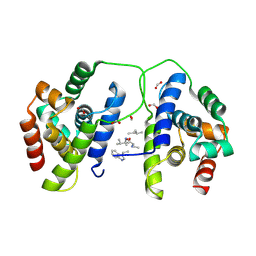 | |
8BRH
 
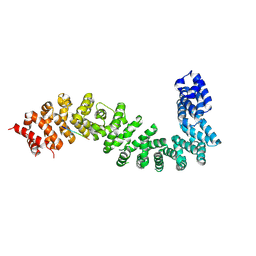 | | Co-crystal structure of She4 with Myo4 peptide | | Descriptor: | KLLA0E16699p, Myo4 peptide (LYS-PHE-ILE-VAL-SER-HIS-TYR) | | Authors: | Arnese, R, Gudino, R, Meinhart, A, Clausen, T. | | Deposit date: | 2022-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UNC-45 assisted myosin folding depends on a conserved FX 3 HY motif implicated in Freeman Sheldon Syndrome.
Nat Commun, 15, 2024
|
|
8BRG
 
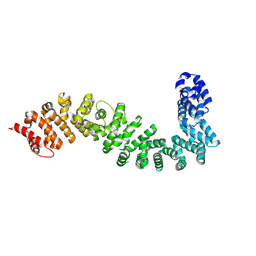 | | Crystal structure of She4 | | Descriptor: | KLLA0E16699p | | Authors: | Gudino, R, Arnese, R, Meinhart, A, Clausen, T. | | Deposit date: | 2022-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UNC-45 assisted myosin folding depends on a conserved FX 3 HY motif implicated in Freeman Sheldon Syndrome.
Nat Commun, 15, 2024
|
|
4XW3
 
 | |
6EPH
 
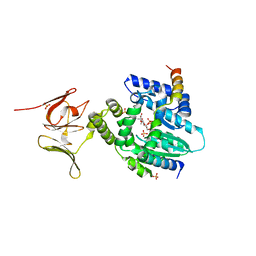 | | Structure of the epsilon_1 / zeta_1 antitoxin toxin system from Neisseria gonorrhoeae in complex with UNAM. | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, CITRATE ANION, Epsilon_1 antitoxin, ... | | Authors: | Rocker, A, Meinhart, A. | | Deposit date: | 2017-10-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The ng_ zeta 1 toxin of the gonococcal epsilon/zeta toxin/antitoxin system drains precursors for cell wall synthesis.
Nat Commun, 9, 2018
|
|
6EPG
 
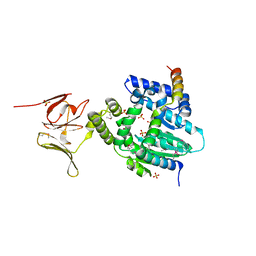 | |
6TV6
 
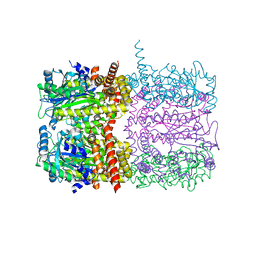 | | Octameric McsB from Bacillus subtilis. | | Descriptor: | MAGNESIUM ION, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Hajdusits, B, Meinhart, A, Clausen, T. | | Deposit date: | 2020-01-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation.
Elife, 10, 2021
|
|
6EPI
 
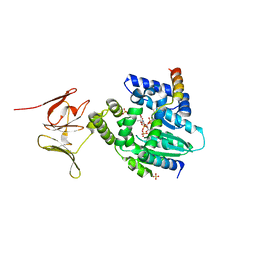 | |
3ISM
 
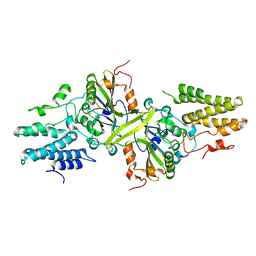 | | Crystal structure of the EndoG/EndoGI complex: Mechanism of EndoG inhibition | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CG4930, CG8862, ... | | Authors: | Loll, B, Gebhardt, M, Wahle, E, Meinhart, A. | | Deposit date: | 2009-08-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the EndoG/EndoGI complex: mechanism of EndoG inhibition.
Nucleic Acids Res., 37, 2009
|
|
3Q8X
 
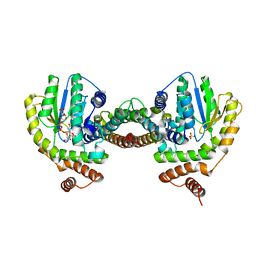 | | Structure of a toxin-antitoxin system bound to its substrate | | Descriptor: | Antidote of epsilon-zeta postsegregational killing system, SULFATE ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, ... | | Authors: | Mutschler, H, Gebhardt, M, Shoeman, R.L, Meinhart, A. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis.
Plos Biol., 9, 2011
|
|
5FGT
 
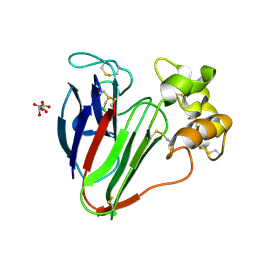 | | Thaumatin solved by native sulphur-SAD using free-electron laser radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.K, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
5FGX
 
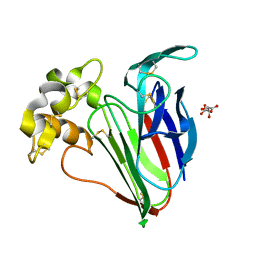 | | Thaumatin solved by native sulphur SAD using synchrotron radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.J, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
2KM4
 
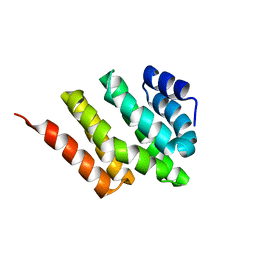 | | Solution structure of Rtt103 CTD interacting domain | | Descriptor: | Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S, Kim, M, Leeper, T.C, Becker, R, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2009-07-20 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6TAY
 
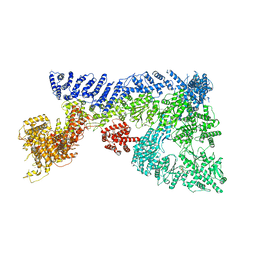 | | Mouse RNF213 mutant R4753K modeling the Moyamoya-disease-related Human variant R4810K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
