7ZRA
 
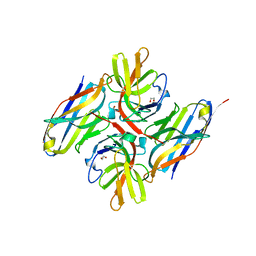 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS1(Nb14497) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody NbSOS1 (Nb14497) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2022-05-04 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
7OCJ
 
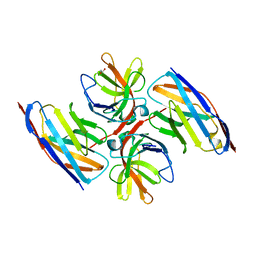 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
7B5G
 
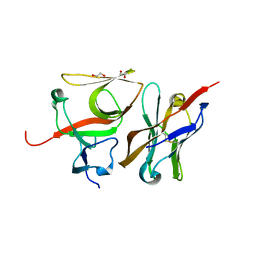 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS3(Nb14527) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody Nb14527, ... | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2020-12-03 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
7AAI
 
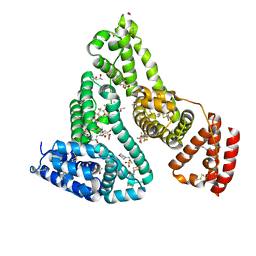 | | Crystal structure of Human serum albumin in complex with perfluorooctanoic acid (PFOA) at 2.10 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
7AAE
 
 | | Crystal structure of Human serum albumin in complex with myristic acid at 2.27 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Albumin, FORMIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
8VRB
 
 | | Structure of a synthetic antibody in complex with a class I MHC presenting a hapten-peptide conjugate | | Descriptor: | AMG 510 (bound form), Beta-2-microglobulin, GTPase KRas, ... | | Authors: | Maso, L, Bang, I, Koide, S. | | Deposit date: | 2024-01-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular basis for antibody recognition of multiple drug-peptide/MHC complexes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VR9
 
 | | Structure of a synthetic antibody in complex with a class I MHC presenting a hapten-peptide conjugate | | Descriptor: | AMG 510 (bound form), Beta-2-microglobulin, GTPase KRas, ... | | Authors: | Maso, L, Bang, I, Koide, S. | | Deposit date: | 2024-01-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Molecular basis for antibody recognition of multiple drug-peptide/MHC complexes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VRA
 
 | | Structure of a synthetic antibody in complex with a class I MHC presenting a hapten-peptide conjugate | | Descriptor: | AMG 510 (bound form), Beta-2-microglobulin, GTPase KRas, ... | | Authors: | Maso, L, Bang, I, Koide, S. | | Deposit date: | 2024-01-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Molecular basis for antibody recognition of multiple drug-peptide/MHC complexes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6IBV
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd 1 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6IBS
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 6 | | Descriptor: | CALCIUM ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6Q35
 
 | | Crystal structure of GES-5 beta-lactamase in complex with boronic inhibitor cpd 3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6Q30
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 5 | | Descriptor: | (7-carboxy-1-benzothiophen-2-yl)-tris(oxidanyl)boranuide, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6Q2Y
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6TGI
 
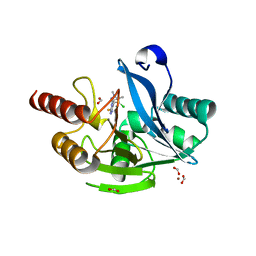 | | Crystal structure of VIM-2 in complex with triazole-based inhibitor OP24 | | Descriptor: | 5-(4-chloranyl-1,5-dimethyl-pyrazol-3-yl)-4-ethyl-1,2,4-triazole-3-thiol, FORMIC ACID, Vim-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
6TGD
 
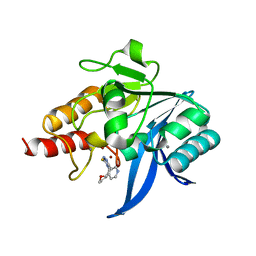 | | Crystal structure of NDM-1 in complex with triazole-based inhibitor OP31 | | Descriptor: | 4-[[(2~{S})-oxolan-2-yl]methyl]-3-pyridin-3-yl-1~{H}-1,2,4-triazole-5-thione, CALCIUM ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
6TS9
 
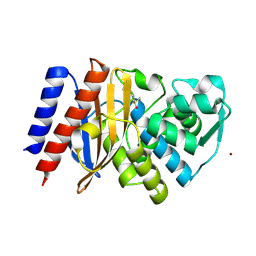 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-lactamase, ... | | Authors: | Maso, L, Tondi, D, Klein, R, Montanari, M, Bellio, C, Celenza, G, Brenk, R, Cendron, L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the Class A Carbapenemase GES-5 via Virtual Screening.
Biomolecules, 10, 2020
|
|
8B0V
 
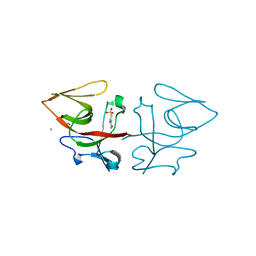 | | Crystal structure of C-terminal domain of Pseudomonas aeruginosa LexA G91D mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Vascon, F, De Felice, S, Chinellato, M, Maso, L, Cendron, L. | | Deposit date: | 2022-09-08 | | Release date: | 2024-03-27 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of Pseudomonas aeruginosa SOS response reveal structural requisites for LexA autoproteolysis.
Iscience, 28, 2025
|
|
4NUI
 
 | | Crystal structure of cobalt-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, COBALT (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUN
 
 | | Crystal structure of copper-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, COPPER (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUO
 
 | | Crystal structure of zinc-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, ZINC ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUK
 
 | | Crystal structure of nickel-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, NICKEL (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
8S70
 
 | | Cryo-EM structure of Pseudomonas aeruginosa recombinase A (RecA) in complex with ssDNA 72mer and ATPgS | | Descriptor: | DNA, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | De Felice, S, Vascon, F, Huber, S.T, Grinzato, A, Jakobi, A.J, Cendron, L. | | Deposit date: | 2024-02-28 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Snapshots of Pseudomonas aeruginosa SOS response reveal structural requisites for LexA autoproteolysis.
Iscience, 28, 2025
|
|
8S7G
 
 | | Cryo-EM structure of Pseudomonas aeruginosa Recombinase A (RecA) in complex with LexAS125A mutant | | Descriptor: | DNA (36-MER), LexA repressor, MAGNESIUM ION, ... | | Authors: | De Felice, S, Vascon, F, Huber, S.T, Catalano, C, Jakobi, A.J, Cendron, L. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Snapshots of Pseudomonas aeruginosa SOS response reveal structural requisites for LexA autoproteolysis.
Iscience, 28, 2025
|
|
8EZG
 
 | | Monobody 12D1 bound to KRAS(G12D) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-10-31 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F0M
 
 | | Monobody 12D5 bound to KRAS(G12D) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Hattori, T, Glasser, E, Akkapeddi, P, Ketavarapu, G, Teng, K.W, Koide, A, Koide, S. | | Deposit date: | 2022-11-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Exploring switch II pocket conformation of KRAS(G12D) with mutant-selective monobody inhibitors.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
