6XA2
 
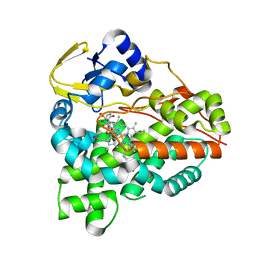 | | Structure of the tirandamycin C-bound P450 monooxygenase TamI | | Descriptor: | (3E)-3-{(2E,4E,6R)-1-hydroxy-4-methyl-6-[(1R,3R,4S,5R)-1,4,8-trimethyl-2,9-dioxabicyclo[3.3.1]non-7-en-3-yl]hepta-2,4-dien-1-ylidene}-2H-pyrrole-2,4(3H)-dione, PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
6XA3
 
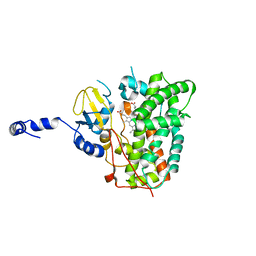 | | Structure of the ligand free P450 monooxygenase TamI | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
5KCI
 
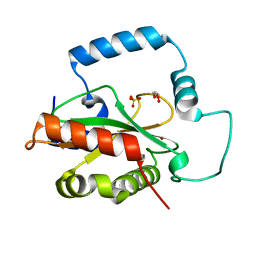 | | Crystal Structure of HTC1 | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein YPL067C, ... | | Authors: | Martin, R.M, Horowitz, S, Koepnick, B, Cooper, S, Flatten, J, Rogawski, D.S, Koropatkin, N.M, Beinlich, F.R.M, Players, F, Students, U.M, Popovic, Z, Baker, D, Khatib, F, Bardwell, J.C.A. | | Deposit date: | 2016-06-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Determining crystal structures through crowdsourcing and coursework.
Nat Commun, 7, 2016
|
|
4UFQ
 
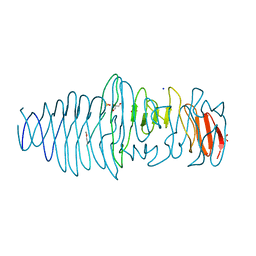 | | Structure of a novel Hyaluronidase (Hyal_Sk) from Streptomyces koganeiensis. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gavira, J.A, Messina, L, Pernagallo, S, Unciti-Broceta, J.D, Conejero-Muriel, M, Diaz-Mochon, J.J, Vaccaro, S, Caruso, S, Musumeci, L, Bisicchia, S, Di Pasquale, R. | | Deposit date: | 2015-03-18 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification and Characterization of a Bacterial Hyaluronidase and its Production in Recombinant Form.
FEBS Lett., 590, 2016
|
|
8A7O
 
 | |
8A7T
 
 | |
8A7P
 
 | |
8A7Q
 
 | |
7NMO
 
 | | Crystal structure of beta-2-microglobulin D76A mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMR
 
 | | Crystal structure of beta-2-microglobulin D76S mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMT
 
 | | Crystal structure of beta-2-microglobulin D76G mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMC
 
 | | Crystal structure of beta-2-microglobulin D76E mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL, TRIETHYLENE GLYCOL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMV
 
 | | Crystal structure of beta-2-microglobulin D76Q mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMY
 
 | | Crystal structure of beta-2-microglobulin D76Y mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NN5
 
 | | Crystal structure of beta-2-microglobulin D76K mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-24 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OY5
 
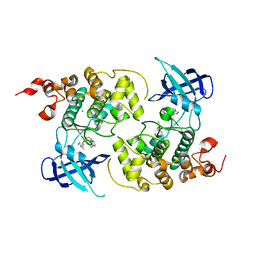 | | Crystal structure of GSK3Beta in complex with ARN25068 | | Descriptor: | CHLORIDE ION, Glycogen synthase kinase-3 beta, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Giabbai, B, Storici, P, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
7OY6
 
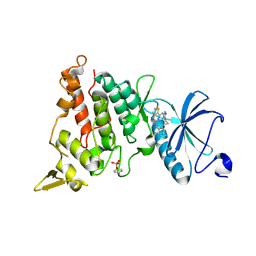 | | Crystal structure of human DYRK1A in complex with ARN25068 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
6HK4
 
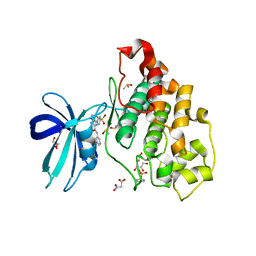 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C22 | | Descriptor: | 3-azanyl-6-(4-morpholin-4-ylsulfonylphenyl)-~{N}-pyridin-3-yl-pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLY-SER-HIS-GLY-HIS-HIS-HIS-HIS-HIS, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6HK3
 
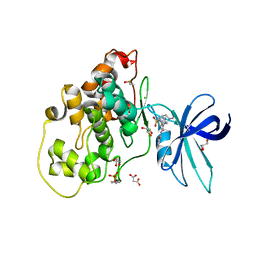 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C44 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyphenyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6HK7
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C50 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyethyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
7Z2P
 
 | | Tubulin-nocodazole complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Prota, A.E, Muehlethaler, T, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
7Z2N
 
 | | Tubulin-18-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
