7DSE
 
 | | CALHM1 close state with ordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
1HPZ
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, POL POLYPROTEIN | | Authors: | Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
7WH9
 
 | | holo structure of emodin 1-OH O-methyltransferase complex with emodin and S-Adenosyl-L-homocysteine | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, O-methyltransferase gedA, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liang, Y.J, Lu, X.F, Qi, F.F, Xue, Y.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Characterization and Structural Analysis of Emodin- O -Methyltransferase from Aspergillus terreus.
J.Agric.Food Chem., 70, 2022
|
|
4IRV
 
 | |
4JSO
 
 | |
4JSD
 
 | |
8OZE
 
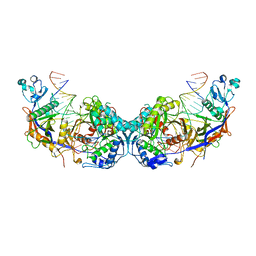 | | cryoEM structure of SPARTA complex dimer high resolution | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*C)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*G)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZ6
 
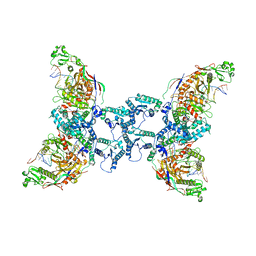 | | cryoEM structure of SPARTA complex ligand-free | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZF
 
 | | cryoEM structure of SPARTA complex Tetramer Post-NAD cleavage-2 | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZG
 
 | | cryoEM structure of SPARTA complex Tetramer Post-NAD cleavage-1 | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZI
 
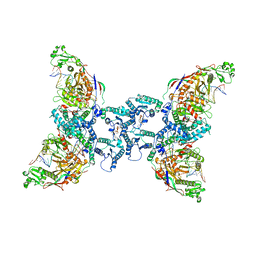 | | cryoEM structure of SPARTA complex pre-NAD cleavage | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain-containing protein, ... | | Authors: | Babatunde, E, Dong, C.N, Xu, H.L, Henning, S. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
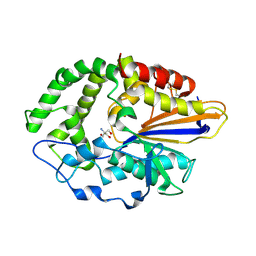
4JOD
 
 | |
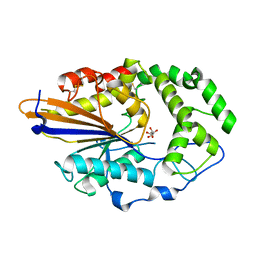
4JOB
 
 | |
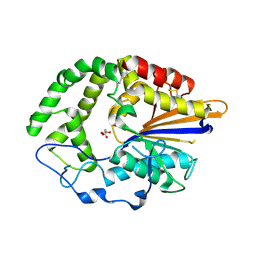
4JOC
 
 | |
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
6HP9
 
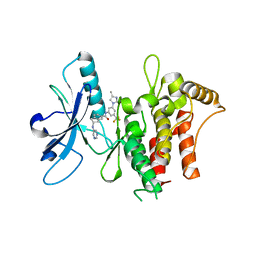 | | Structure of the kinase domain of human DDR1 in complex with a 2-Amino-2,3-Dihydro-1H-Indene-5-Carboxamide-based inhibitor | | Descriptor: | (2~{R})-~{N}-[3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)phenyl]-2-(pyrimidin-5-ylamino)-2,3-dihydro-1~{H}-indene-5-carboxamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Amino-2,3-dihydro-1H-indene-5-carboxamide-Based Discoidin Domain Receptor 1 (DDR1) Inhibitors: Design, Synthesis, and in Vivo Antipancreatic Cancer Efficacy.
J.Med.Chem., 62, 2019
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
6JRK
 
 | | The structure of co-crystals of 8r-B-EGFR WT complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6JRJ
 
 | | The structure of co-crystals of 8r-B-EGFR T790M/C797S complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.943 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6JRX
 
 | | EGFR T790M/C797S in complex with compound 6i | | Descriptor: | Epidermal growth factor receptor, N-{trans-4-[3-(2-chlorophenyl)-7-{[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]cyclohexyl}propanamide | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JUU
 
 | | Crystal structure of ZAK in complex with compound 6r | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]naphthalene-1-sulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JUT
 
 | | Crystal structure of ZAK in complex with compound 6k | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]-3-bromanyl-benzenesulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
4NSQ
 
 | | Crystal structure of PCAF | | Descriptor: | COENZYME A, Histone acetyltransferase KAT2B | | Authors: | Lin, J.Y, Cai, Y.F. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3108 Å) | | Cite: | Dimeric structure of p300/CBP associated factor.
Bmc Struct.Biol., 14, 2014
|
|
3CW9
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase in the Thioester-forming Conformation, bound to 4-chlorophenacyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-Chlorophenacyl-coenzyme A, 4-chlorobenzoyl CoA ligase, ... | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
