6U2P
 
 | | PCSK9 in complex with compound 5 | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoro-4-{[(1R)-6-methoxy-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
4DXD
 
 | | Staphylococcal Aureus FtsZ in complex with 723 | | Descriptor: | 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lu, J, Soisson, S.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Restoring methicillin-resistant Staphylococcus aureus susceptibility to beta-lactam antibiotics.
Sci Transl Med, 4, 2012
|
|
3D8A
 
 | | Co-crystal structure of TraM-TraD complex. | | Descriptor: | Protein traD, Relaxosome protein TraM | | Authors: | Glover, J.N.M, Lu, J, Wong, J.J, Edwards, R.A. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation.
Mol.Microbiol., 70, 2008
|
|
1I3Z
 
 | | MURINE EAT2 SH2 DOMAIN IN COMPLEX WITH SLAM PHOSPHOPEPTIDE | | Descriptor: | EWS/FLI1 ACTIVATED TRANSCRIPT 2, SIGNALING LYMPHOCYTIC ACTIVATION MOLECULE | | Authors: | Lu, J, Poy, F, Morra, M, Terhorst, C, Eck, M.J. | | Deposit date: | 2001-02-19 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the interaction of the free SH2 domain EAT-2 with
SLAM receptors in hematopoietic cells.
Eur.J.Biochem., 20, 2001
|
|
6CYB
 
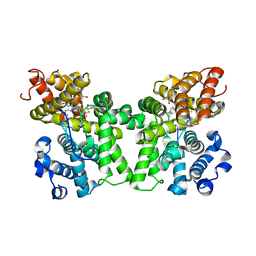 | | PDE2 in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2,2,2-trifluoroethyl)-1-{(1S)-1-[4-(trifluoromethyl)phenyl]ethyl}-1H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design and Procognitive Assessment of a Potent and Selective Phosphodiesterase 2A Inhibitor.
ACS Med Chem Lett, 9, 2018
|
|
6CYD
 
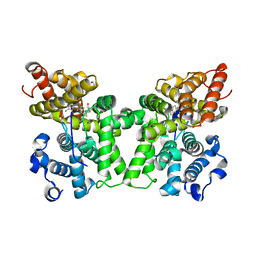 | | PDE2 in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 3-(hydroxymethyl)-6-methyl-1-{(1S)-1-[4-(trifluoromethyl)phenyl]ethyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Guided Design and Procognitive Assessment of a Potent and Selective Phosphodiesterase 2A Inhibitor.
ACS Med Chem Lett, 9, 2018
|
|
8DKJ
 
 | |
8DMN
 
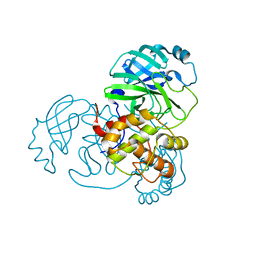 | |
8DI3
 
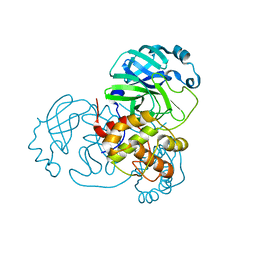 | |
8DKL
 
 | |
8DKK
 
 | |
8DKH
 
 | |
8EJ9
 
 | |
8EJ7
 
 | |
5BS3
 
 | | Crystal Structure of S.A. gyrase in complex with Compound 7 | | Descriptor: | (4R)-3-fluoro-4-hydroxy-4-{[(1r,4R)-4-{[(3-oxo-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazin-6-yl)methyl]amino}-2-oxabicyclo[2.2.2]oct-1-yl]methyl}-4,5-dihydro-7H-pyrrolo[3,2,1-de][1,5]naphthyridin-7-one, DNA gyrase subunit A and B, DNA/RNA (5'-R(P*AP*GP*CP*CP*G)-D(P*T)-R(P*AP*GP*GP*GP*CP*CP*C)-D(P*T)-R(P*AP*CP*GP*GP*C)-D(P*T)-3'), ... | | Authors: | Lu, J, Patel, S, Soisson, S. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tricyclic 1,5-naphthyridinone oxabicyclooctane-linked novel bacterial topoisomerase inhibitors as broad-spectrum antibacterial agents-SAR of left-hand-side moiety (Part-2).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7CFU
 
 | |
4PLB
 
 | | Crystal Structure of S.A. gyrase-AM8191 complex | | Descriptor: | 6-[({(1r,4S)-1-[(1S)-2-(3-fluoro-6-methoxy-1,5-naphthyridin-4-yl)-1-hydroxyethyl]-2-oxabicyclo[2.2.2]oct-4-yl}amino)methyl]-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, Chimera protein of DNA gyrase subunits B and A, DNA (5'-D(P*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Lu, J, Patel, S, Soisson, S. | | Deposit date: | 2014-05-16 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Oxabicyclooctane-linked novel bacterial topoisomerase inhibitors as broad spectrum antibacterial agents.
Acs Med.Chem.Lett., 5, 2014
|
|
8DUW
 
 | | HnRNPA2 D290V LCD PM2 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
8DU2
 
 | | HnRNPA2 D290V LCD PM1 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
6B97
 
 | | Crystal structure of PDE2 in complex with complex 9 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-1-methyl-N-{(1R)-1-[4-(trifluoromethyl)phenyl]ethyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The identification of a novel lead class for phosphodiesterase 2 inhibition by fragment-based drug design.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6B98
 
 | | PDE2 in complex with compound 1 | | Descriptor: | 6-chloro-N,1-dimethyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Lu, J. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The identification of a novel lead class for phosphodiesterase 2 inhibition by fragment-based drug design.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6B96
 
 | | Crystal Structure of PDE2 in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-N-{1-[4-(trifluoromethyl)phenyl]cyclopropyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The identification of a novel lead class for phosphodiesterase 2 inhibition by fragment-based drug design.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2JSZ
 
 | | Solution structure of Tpx in the reduced state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J, Yang, F. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2JSY
 
 | | Solution structure of Tpx in the oxidized state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
6CYC
 
 | | PDE2 in complex with compound 5 | | Descriptor: | 1,2-ETHANEDIOL, 3-(hydroxymethyl)-1-{(1S)-1-[4-(trifluoromethyl)phenyl]ethyl}-1H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-Guided Design and Procognitive Assessment of a Potent and Selective Phosphodiesterase 2A Inhibitor.
ACS Med Chem Lett, 9, 2018
|
|
