1RKB
 
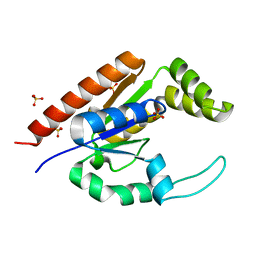 | | The structure of adrenal gland protein AD-004 | | Descriptor: | LITHIUM ION, Protein AD-004, SULFATE ION | | Authors: | Ren, H, Liang, Y, Bennett, M, Su, X.D. | | Deposit date: | 2003-11-21 | | Release date: | 2005-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human adenylate kinase 6: An adenylate kinase localized to the cell nucleus
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7LVM
 
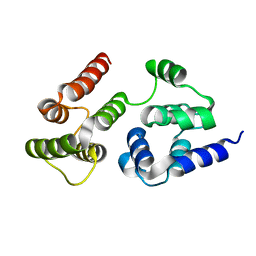 | |
7LVJ
 
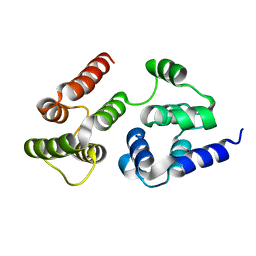 | | CASP8 isoform G DED domain | | Descriptor: | Isoform 9 of Caspase-8 | | Authors: | Weichert, K, Lu, F, Kodandapani, L, Sauder, J.M. | | Deposit date: | 2021-02-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caspase-8 Variant G Regulates Rheumatoid Arthritis Fibroblast-Like Synoviocyte Aggressive Behavior.
ACR Open Rheumatol, 4, 2022
|
|
5J40
 
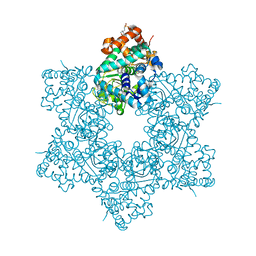 | | The X-ray structure of JCV Helicase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Large T antigen, SULFATE ION, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment-Based Discovery of Dual JC Virus and BK Virus Helicase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5J4Y
 
 | |
5J47
 
 | | The X-ray structure of Inhibitor Bound to JCV Helicase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-phenoxyphenyl)[1,2,4]triazolo[3,4-b][1,3,4]thiadiazole, Large T antigen, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Dual JC Virus and BK Virus Helicase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5J4V
 
 | | The crystal structure of Inhibitor Bound to JCV Helicase | | Descriptor: | 2-(2-phenoxypyridin-3-yl)[1,3]thiazolo[5,4-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Large T antigen, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Fragment-Based Discovery of Dual JC Virus and BK Virus Helicase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4K2J
 
 | |
4PGG
 
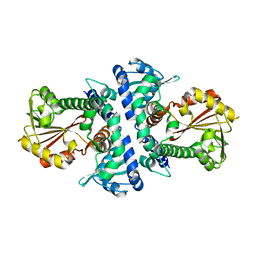 | | Caffeic acid O-methyltransferase from Sorghum bicolor | | Descriptor: | Caffeic acid O-methyltransferase | | Authors: | Green, A.R, Lewis, K.M, Kang, C. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Determination of the Structure and Catalytic Mechanism of Sorghum bicolor Caffeic Acid O-Methyltransferase and the Structural Impact of Three brown midrib12 Mutations.
Plant Physiol., 165, 2014
|
|
4PGH
 
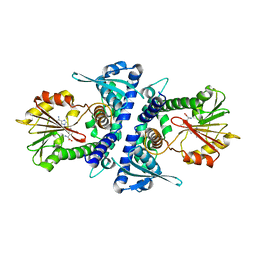 | | Caffeic acid O-methyltransferase from Sorghum bicolor | | Descriptor: | Caffeic acid O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Green, A.R, Lewis, K.M, Kang, C. | | Deposit date: | 2014-05-02 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Determination of the Structure and Catalytic Mechanism of Sorghum bicolor Caffeic Acid O-Methyltransferase and the Structural Impact of Three brown midrib12 Mutations.
Plant Physiol., 165, 2014
|
|
2EXX
 
 | | Crystal structure of HSCARG from Homo sapiens in complex with NADP | | Descriptor: | GLYCEROL, HSCARG protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dai, X, Chen, Q, Yao, D, Liang, Y, Dong, Y, Gu, X, Zheng, X, Luo, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5CNK
 
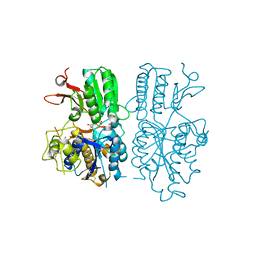 | | mglur3 with glutamate | | Descriptor: | GLUTAMIC ACID, IODIDE ION, Metabotropic glutamate receptor 3 | | Authors: | Monn, J.A, Clawson, D.K. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
5CNJ
 
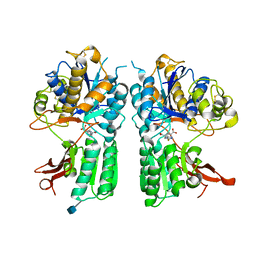 | | mGlur2 with glutamate analog | | Descriptor: | (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Monn, J.A, Clawson, D.K. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
5CNI
 
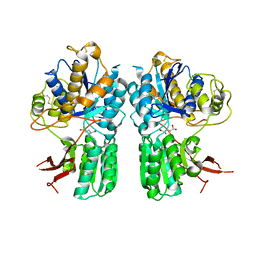 | | mGlu2 with Glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Clawson, D.K, Atwell, S, Monn, J.A. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
5CNM
 
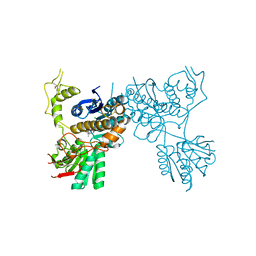 | | mGluR3 complexed with glutamate analog | | Descriptor: | (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Monn, J.A, Clawson, D.K, McKinzie, D. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
7SFT
 
 | | Filamin complex-2 | | Descriptor: | Filamin-A, Integrin alpha-IIb light chain, form 2 | | Authors: | Liu, J, Qin, J. | | Deposit date: | 2021-10-04 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | A mechanism of platelet integrin alpha IIb beta 3 outside-in signaling through a novel integrin alpha IIb subunit-filamin-actin linkage.
Blood, 141, 2023
|
|
7SC4
 
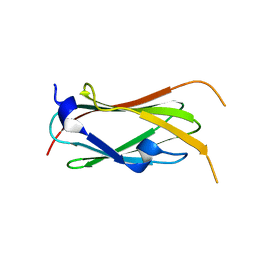 | | filamin complex-1 | | Descriptor: | Filamin-A/Integrin alpha-IIb light chain, form 2 chimera, SULFATE ION | | Authors: | Liu, J, Qin, J. | | Deposit date: | 2021-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A mechanism of platelet integrin alpha IIb beta 3 outside-in signaling through a novel integrin alpha IIb subunit-filamin-actin linkage.
Blood, 141, 2023
|
|
6B7H
 
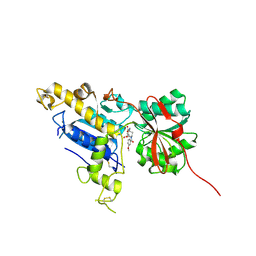 | | Structure of mGluR3 with an agonist | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Monn, J.A, Clawson, D.K. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4beta-Amide-Substituted 2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1 S,2 S,4 S,5 R,6 S)-2-Amino-4-[(3-methoxybenzoyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2794193), a Highly Potent and Selective mGlu3Receptor Agonist.
J. Med. Chem., 61, 2018
|
|
6BA6
 
 | | Solution structure of Rap1b/talin complex | | Descriptor: | Ras-related protein Rap-1b, Talin-1 | | Authors: | Zhu, L, Yang, J, Qin, J. | | Deposit date: | 2017-10-12 | | Release date: | 2017-12-06 | | Last modified: | 2019-12-04 | | Method: | SOLUTION NMR | | Cite: | Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation.
Nat Commun, 8, 2017
|
|
5VB9
 
 | | IL-17A in complex with peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-17A, ... | | Authors: | Antonysamy, S, Russell, M, Zhang, A, Groshong, C, Manglicmot, D, Lu, F, Benach, J, Wasserman, S.R, Zhang, F, Afshar, S, Bina, H, Broughton, H, Chalmers, M, Dodge, J, Espada, A, Jones, S, Ting, J.P, Woodman, M. | | Deposit date: | 2017-03-28 | | Release date: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Utilization of peptide phage display to investigate hotspots on IL-17A and what it means for drug discovery.
PLoS ONE, 13, 2018
|
|
5U73
 
 | | Crystal structure of human Niemann-Pick C1 protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Niemann-Pick C1 protein, ... | | Authors: | Li, X, Wang, J, Blobel, G. | | Deposit date: | 2016-12-11 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U74
 
 | | Structure of human Niemann-Pick C1 protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6JQ9
 
 | | Crystal structure of a lyase from Alteromonas sp. | | Descriptor: | CALCIUM ION, SULFATE ION, Short ulvan lyase | | Authors: | Qin, H.M, Guo, Q.Q. | | Deposit date: | 2019-03-29 | | Release date: | 2020-04-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Biochemical characterization and structural analysis of ulvan lyase from marine Alteromonas sp. reveals the basis for its salt tolerance.
Int.J.Biol.Macromol., 147, 2020
|
|
6JNL
 
 | | REF6 ZnF2-4-NAC004 complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
6JQX
 
 | |
