2RHO
 
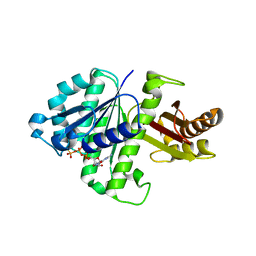 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ NCS Dimer with Bound GDP and GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell Division Protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
2RHL
 
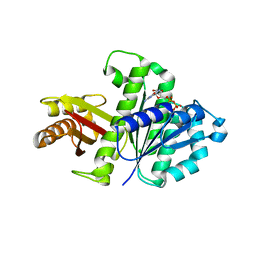 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ NCS Dimer with Bound GDP | | Descriptor: | Cell Division Protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
2RHH
 
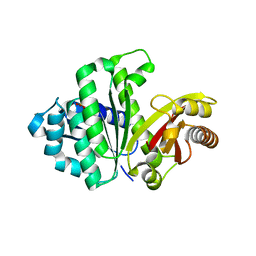 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ with Bound Sulfate Ion | | Descriptor: | Cell Division Protein ftsZ, SULFATE ION | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | Descriptor: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
6NKL
 
 | | 2.2 A resolution structure of VapBC-1 from nontypeable Haemophilus influenzae | | Descriptor: | Antitoxin VapB1, Ribonuclease VapC | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Molinaro, A.L, Daines, D.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of VapBC-1 from Nontypeable Haemophilus influenzae and the Effect of PIN Domain Mutations on Survival during Infection.
J.Bacteriol., 201, 2019
|
|
6UJI
 
 | |
6VGY
 
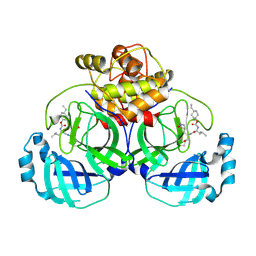 | | 2.05 A resolution structure of MERS 3CL protease in complex with inhibitor 6b | | Descriptor: | N~2~-{[(trans-4-ethylcyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH3
 
 | | 2.20 A resolution structure of MERS 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH2
 
 | | 2.26 A resolution structure of MERS 3CL protease in complex with inhibitor 7i | | Descriptor: | 4,4-difluorocyclohexyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH0
 
 | | 1.95 A resolution structure of MERS 3CL protease in complex with inhibitor 6g | | Descriptor: | N~2~-{[(5-ethyl-1,3-dioxan-5-yl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH1
 
 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | Descriptor: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VGZ
 
 | | 2.25 A resolution structure of MERS 3CL protease in complex with inhibitor 6d | | Descriptor: | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
3R9I
 
 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
3UR9
 
 | | 1.65A resolution structure of Norwalk Virus Protease Containing a covalently bound dipeptidyl inhibitor | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like protease, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
3UR6
 
 | | 1.5A resolution structure of apo Norwalk Virus Protease | | Descriptor: | 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
5HKW
 
 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
5HKY
 
 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
5HKZ
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
5HL0
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
5HKX
 
 | | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CBL, SODIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution
To be published
|
|
5IXE
 
 | | 1.75A RESOLUTION STRUCTURE OF 5-Fluoroindole BOUND BETA-GLYCOSIDASE (W33G) FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-fluoro-1H-indole, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Full and Partial Agonism of a Designed Enzyme Switch.
ACS Synth Biol, 5, 2016
|
|
8EJX
 
 | |
4DCD
 
 | | 1.6A resolution structure of PolioVirus 3C Protease Containing a covalently bound dipeptidyl inhibitor | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Genome polyprotein, ... | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.-C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2012-01-17 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
4DM0
 
 | | TN5 transposase: 20MER OUTSIDE END 2 MN complex | | Descriptor: | 1,2-ETHANEDIOL, DNA NON-TRANSFERRED STRAND, DNA TRANSFERRED STRAND, ... | | Authors: | Klenchin, V.A, Lovell, S, Goryshin, I.Y, Reznikoff, W.R, Rayment, I. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-metal active site binding of a TN5 transposase synaptic complex
Nat.Struct.Biol., 9, 2002
|
|
4E6K
 
 | | 2.0 A resolution structure of Pseudomonas aeruginosa bacterioferritin (BfrB) in complex with bacterioferritin associated ferredoxin (Bfd) | | Descriptor: | Bacterioferritin, FE2/S2 (INORGANIC) CLUSTER, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Wang, Y, Kumar, R, Ruvinsky, A, Vasker, I, Rivera, M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
J.Am.Chem.Soc., 134, 2012
|
|
