4ZS7
 
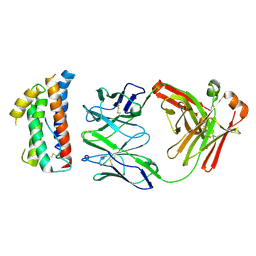 | | Structural mimicry of receptor interaction by antagonistic IL-6 antibodies | | Descriptor: | Interleukin-6, Llama Fab fragment 68F2 heavy chain, Llama Fab fragment 68F2 light chain | | Authors: | Blanchetot, C, De Jonge, N, Desmyter, A, Ongenae, N, Hofman, E, Klarenbeek, A, Sadi, A, Hultberg, A, Kretz-Rommel, A, Spinelli, S, Loris, R, Cambillau, C, de Haard, H. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
J.Biol.Chem., 291, 2016
|
|
5HOH
 
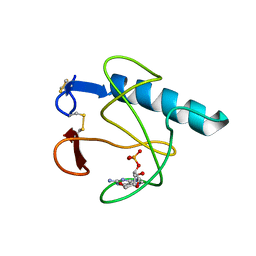 | | RIBONUCLEASE T1 (ASN9ALA/THR93ALA DOUBLEMUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
5J9I
 
 | |
5JA9
 
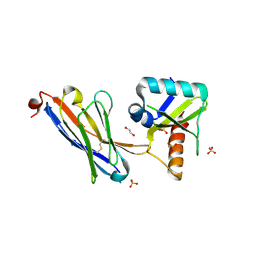 | | Crystal structure of the HigB2 toxin in complex with Nb6 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody 6, SULFATE ION, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-05 | | Last modified: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
5JAA
 
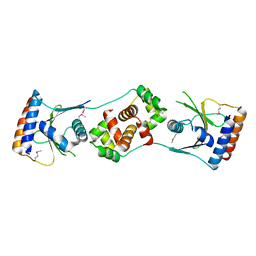 | |
5JA8
 
 | | Crystal structure of the HigB2 toxin in complex with Nb2 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-05 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
1X75
 
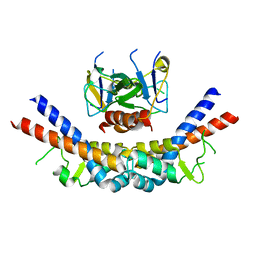 | | CcdB:GyrA14 complex | | Descriptor: | Cytotoxic protein ccdB, DNA gyrase subunit A | | Authors: | Dao-Thi, M.H, Van Melderen, L, De Genst, E, Wyns, L, Loris, R. | | Deposit date: | 2004-08-13 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Basis of Gyrase Poisoning by the Addiction Toxin CcdB
J.Mol.Biol., 348, 2005
|
|
1XFP
 
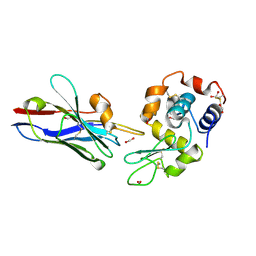 | | Crystal structure of the CDR2 germline reversion mutant of cAb-Lys3 in complex with hen egg white lysozyme | | Descriptor: | FORMIC ACID, Lysozyme C, heavy chain antibody | | Authors: | De Genst, E, Handelberg, F, Van Meirhaeghe, A, Vynck, S, Loris, R, Wyns, L, Muyldermans, S. | | Deposit date: | 2004-09-15 | | Release date: | 2004-09-28 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical Basis for the Affinity Maturation of a Camel Single Domain Antibody
J.Biol.Chem., 279, 2004
|
|
1YC8
 
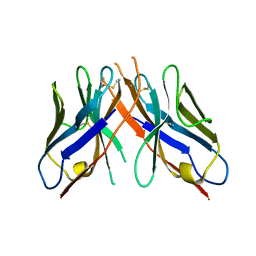 | | cAbAn33- Y37V/E44G/R45L triple mutant | | Descriptor: | anti-VSG immunoglobulin heavy chain variable domain cAbAn33 | | Authors: | Conrath, K, Vincke, C, Stijlemans, B, Schymkowitz, J, Wyns, L, Muyldermans, S, Loris, R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antigen Binding and Solubility Effects upon the Veneering of a Camel VHH in Framework-2 to Mimic a VH.
J.Mol.Biol., 350, 2005
|
|
1YC7
 
 | | cAbAn33 VHH fragment against VSG | | Descriptor: | SULFATE ION, anti-VSG immunoglobulin heavy chain variable domain cAbAn33 | | Authors: | Conrath, K, Vincke, C, Stijlemans, B, Schymkowitz, J, Wyns, L, Muyldermans, S, Loris, R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Antigen Binding and Solubility Effects upon the Veneering of a Camel VHH in Framework-2 to Mimic a VH.
J.Mol.Biol., 350, 2005
|
|
1MMC
 
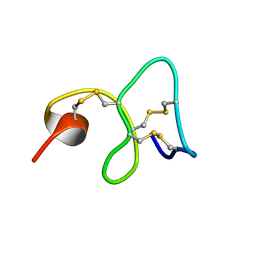 | | 1H NMR STUDY OF THE SOLUTION STRUCTURE OF AC-AMP2 | | Descriptor: | ANTIMICROBIAL PEPTIDE 2 | | Authors: | Martins, J.C, Maes, D, Loris, R, Pepermans, H.A.M, Wyns, L, Willem, R, Verheyden, P. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | H NMR study of the solution structure of Ac-AMP2, a sugar binding antimicrobial protein isolated from Amaranthus caudatus.
J.Mol.Biol., 258, 1996
|
|
1BJQ
 
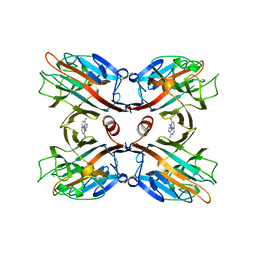 | | THE DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH ADENINE | | Descriptor: | ADENINE, CALCIUM ION, LECTIN, ... | | Authors: | Hamelryck, T.W, Loris, R, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M. | | Deposit date: | 1998-06-26 | | Release date: | 1998-12-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1CES
 
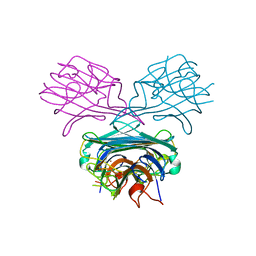 | | CRYSTALS OF DEMETALLIZED CONCANAVALIN A SOAKED WITH ZINC HAVE A ZINC ION BOUND IN THE S1 SITE | | Descriptor: | CONCANAVALIN A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-02-15 | | Release date: | 1997-02-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1IOA
 
 | | ARCELIN-5, A LECTIN-LIKE DEFENSE PROTEIN FROM PHASEOLUS VULGARIS | | Descriptor: | ARCELIN-5A, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hamelryck, T, Loris, R. | | Deposit date: | 1996-10-02 | | Release date: | 1996-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of arcelin-5, a lectin-like defense protein from Phaseolus vulgaris
J.Biol.Chem., 271, 1996
|
|
1ENR
 
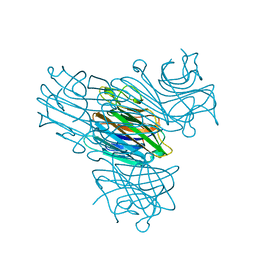 | | CO-CRYSTALS OF DEMETALLIZED CONCANAVALIN A WITH ZINC AND CALCIUM HAVING A ZINC ION BOUND IN THE S1 SITE AND A CALCIUM ION BOUND IN THE S2 SITE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1ENS
 
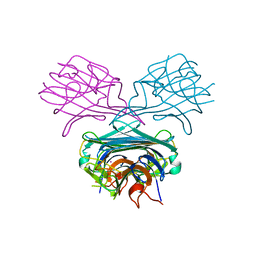 | | CRYSTALS OF DEMETALLIZED CONCANAVALIN A SOAKED WITH COBALT HAVING A COBALT ION BOUND IN THE S1 SITE | | Descriptor: | COBALT (II) ION, CONCANAVALIN A | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1ENQ
 
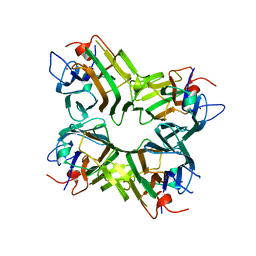 | | CO-CRYSTALS OF DEMETALLIZED CONCANAVALIN A WITH ZINC HAVING A ZINC ION BOUND IN THE S1 SITE | | Descriptor: | CONCANAVALIN A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1G8W
 
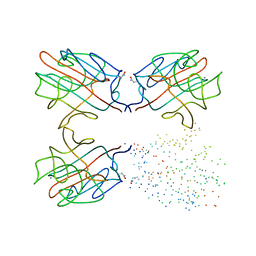 | | IMPROVED STRUCTURE OF PHYTOHEMAGGLUTININ-L FROM THE KIDNEY BEAN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LEUCOAGGLUTINATING PHYTOHEMAGGLUTININ, ... | | Authors: | Buts, L, Hamelryck, T.W, Dao-Thi, M, Loris, R, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-21 | | Release date: | 2000-12-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
1LUL
 
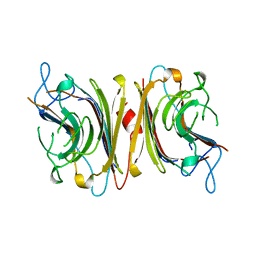 | | DB58, A LEGUME LECTIN FROM DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, LECTIN DB58, MANGANESE (II) ION | | Authors: | Hamelryck, T.W, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M, Loris, R. | | Deposit date: | 1998-06-30 | | Release date: | 1998-12-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
2MF2
 
 | |
3DCQ
 
 | | LECB (PA-LII) in complex with the synthetic ligand 2G0 | | Descriptor: | (2S)-1-[(2S)-6-amino-2-({[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl]acetyl}amino)hexanoyl]-N-[(1S)-1-carbamoyl-3-methylbutyl]pyrrolidine-2-carboxamide, CALCIUM ION, Fucose-binding lectin PA-IIL | | Authors: | Johansson, E.M, Crusz, S.A, Kolomiets, E, Buts, L, Kadam, R.U, Cacciarini, M, Bartels, K.M, Diggle, S.P, Camara, M, Williams, P, Loris, R, Nativi, C, Rosenau, F, Jaeger, K.E, Darbre, T, Reymond, J.L. | | Deposit date: | 2008-06-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB.
Chem.Biol., 15, 2008
|
|
3DD7
 
 | |
3DD9
 
 | | Structure of DocH66Y dimer | | Descriptor: | Death on curing protein | | Authors: | Garcia-Pino, A, Loris, R. | | Deposit date: | 2008-06-05 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The intrinsically disordered domain of the antitoxin Phd chaperones the toxin Doc against irreversible inactivation and misfolding
J. Biol. Chem., 289, 2014
|
|
3DIE
 
 | |
3DWT
 
 | |
