1HVI
 
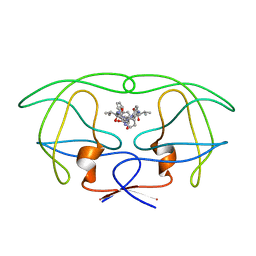 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2R,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
1HVK
 
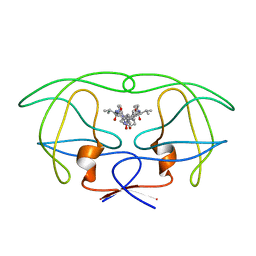 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
1HVL
 
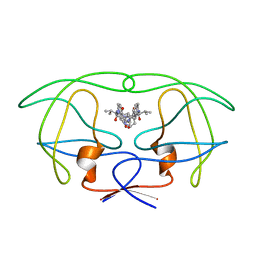 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2R,3R)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
1EER
 
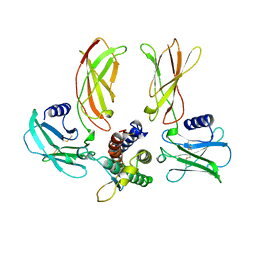 | |
6LNZ
 
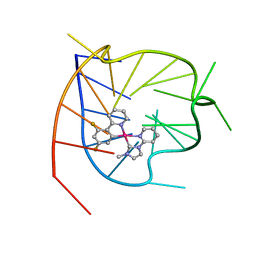 | |
1FSH
 
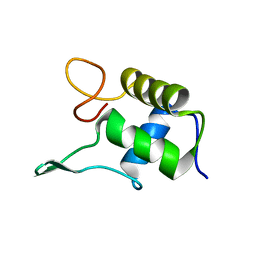 | |
5Y6J
 
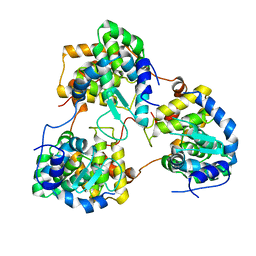 | |
6O1S
 
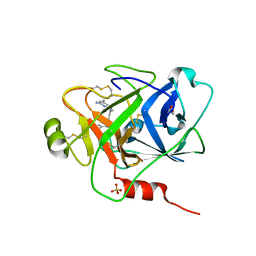 | | Structure of human plasma kallikrein protease domain with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, PHOSPHATE ION, ... | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
5HU4
 
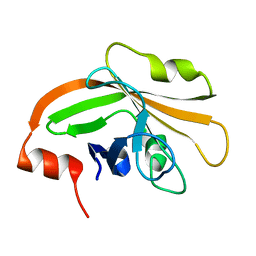 | | Cystal structure of listeria monocytogenes sortase A | | Descriptor: | Cysteine protease | | Authors: | Li, H. | | Deposit date: | 2016-01-27 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of sortase A by chalcone prevents Listeria monocytogenes infection.
Biochem. Pharmacol., 106, 2016
|
|
6JBH
 
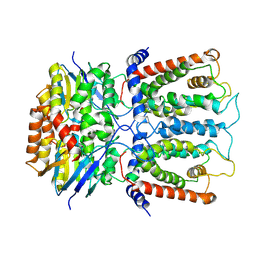 | | Cryo-EM structure and transport mechanism of a wall teichoic acid ABC transporter | | Descriptor: | TarG, TarH | | Authors: | Chen, L, Hou, W.T, Fan, T, Li, Y.H, Liu, B.H, Jiang, Y.L, Sun, L.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter.
Mbio, 11, 2020
|
|
6KHO
 
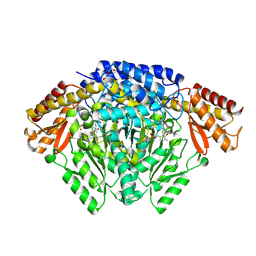 | | Crystal structure of Oryza sativa TDC with PLP | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
6K5M
 
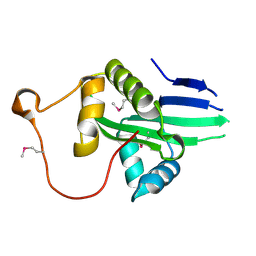 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa (Rice) | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-05-29 | | Release date: | 2020-06-03 | | Last modified: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6KHN
 
 | | Crystal structure of Oryza sativa TDC with PLP and SEROTONIN | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
6KHP
 
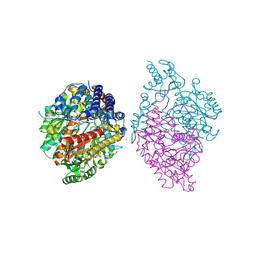 | | Crystal structure of Oryza sativa TDC with PLP and tryptamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, ACETATE ION, CALCIUM ION, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure ofOryza sativaTDC reveals the substrate specificity for TDC-mediated melatonin biosynthesis.
J Adv Res, 24, 2020
|
|
7N5U
 
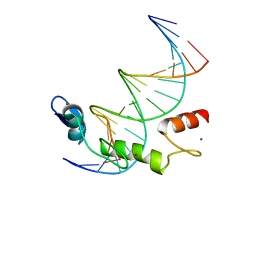 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 21) | | Descriptor: | DNA Strain II, DNA Strand I, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5W
 
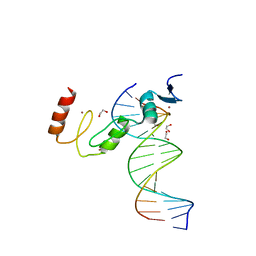 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 23) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand I, DNA Strand II, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5V
 
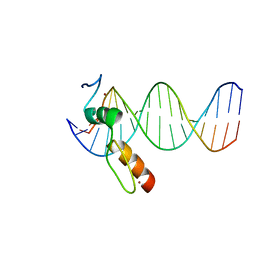 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N9Z
 
 | | E. coli cytochrome bo3 in MSP nanodisc | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vallese, F, Clarke, O.B. | | Deposit date: | 2021-06-19 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo 3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5TJX
 
 | | Structure of human plasma kallikrein | | Descriptor: | (8E)-3-amino-1-methyl-15-[(1H-pyrazol-1-yl)methyl]-7,10,11,12,24,25-hexahydro-6H,18H,23H-19,22-(metheno)pyrido[4,3-j][1,9,13,17,18]benzodioxatriazacyclohenicosin-23-one, PHOSPHATE ION, Plasma kallikrein | | Authors: | Partridge, J.R, Choy, R.M, Li, Z. | | Deposit date: | 2016-10-05 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | Structure-Guided Design of Novel, Potent, and Selective Macrocyclic Plasma Kallikrein Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
8J5J
 
 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
8K8J
 
 | | Cannabinoid Receptor 1 bound to Fenofibrate coupling MiniGsq and Nb35 Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Tang, W.Q, Wang, T.X, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Fenofibrate Recognition and G q Protein Coupling Mechanisms of the Human Cannabinoid Receptor CB1.
Adv Sci, 11, 2024
|
|
