4I2R
 
 | | 2.15 Angstroms X-ray crystal structure of NAD- and alternative substrate-bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-22 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
1QM2
 
 | |
4I1W
 
 | | 2.00 Angstroms X-ray crystal structure of NAD- bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I26
 
 | | 2.20 Angstroms X-ray crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, 2-aminomuconate 6-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Davis, I, Huo, L, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4NPI
 
 | | 1.94 Angstroms X-ray crystal structure of NAD- and intermediate- bound alpha-aminomuconate-epsilon-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2013-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
1QM3
 
 | |
1QLZ
 
 | | Human prion protein | | Descriptor: | PRION PROTEIN | | Authors: | Zahn, R, Liu, A, Luhrs, T, Wuthrich, K. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-16 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4OFC
 
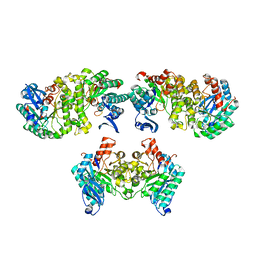 | | 2.0 Angstroms X-ray crystal structure of human 2-amino-3-carboxymuconate-6-semialdehye decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Liu, F, Iwaki, H, Chen, L, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
1QLX
 
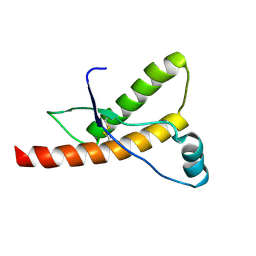 | | Human prion protein | | Descriptor: | PRION PROTEIN | | Authors: | Zahn, R, Liu, A, Luhrs, T, Wuthrich, K. | | Deposit date: | 1999-09-17 | | Release date: | 1999-12-16 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1QM0
 
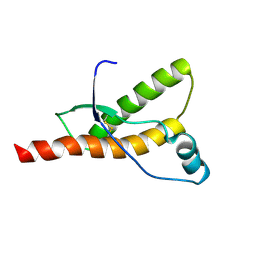 | |
4IH3
 
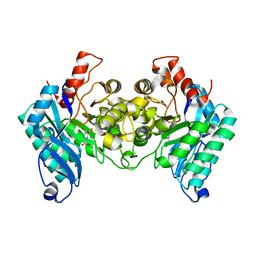 | |
4OE2
 
 | | 2.00 Angstroms X-ray crystal structure of E268A 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Esaki, S, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
2L0K
 
 | | NMR solution structure of a transcription factor SpoIIID in complex with DNA | | Descriptor: | Stage III sporulation protein D | | Authors: | Chen, B, Himes, P, Lu, Z, Liu, A, Yan, H, Kroos, L. | | Deposit date: | 2010-07-08 | | Release date: | 2011-08-17 | | Method: | SOLUTION NMR | | Cite: | Novel Mode of DNA Binding by Bacterial Transcription Factor SpoIIID
To be Published
|
|
5WP2
 
 | | 1.44 Angstrom crystal structure of CYP121 from Mycobacterium tuberculosis in complex with substrate and CN | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, CYANIDE ION, Mycocyclosin synthase, ... | | Authors: | Fielding, A, Dornevil, K, Liu, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Probing Ligand Exchange in the P450 Enzyme CYP121 from Mycobacterium tuberculosis: Dynamic Equilibrium of the Distal Heme Ligand as a Function of pH and Temperature.
J. Am. Chem. Soc., 139, 2017
|
|
3FZ2
 
 | | Crystal structure of the tail terminator protein from phage lambda (gpU-D74A) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
3FZB
 
 | | Crystal structure of the tail terminator protein from phage lambda (gpU-WT) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-24 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
4OUB
 
 | | A 2.20 angstroms X-ray crystal structure of E268A 2-aminomucaonate 6-semialdehyde dehydrogenase catalytic intermediate from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Liu, A. | | Deposit date: | 2014-02-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4OU2
 
 | | A 2.15 Angstroms X-ray crystal structure of E268A 2-aminomuconate 6-semialdehyde dehydrogenase catalytic intermediate from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2,6-dihydroxyhexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Liu, A. | | Deposit date: | 2014-02-14 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4ERG
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, FE (III) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4ERI
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4ERA
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
6BVP
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVQ
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVR
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVS
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.318 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
