6OI9
 
 | | Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(3S)-3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
6OJH
 
 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine, ACETATE ION, Biotin carboxylase, ... | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine
To Be Published
|
|
6OI8
 
 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with 7-((1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl)-6-(2-chloro-6-(pyridin-3-yl)phenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl]-6-[2-chloro-6-(pyridin-3-yl)phenyl]pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
6MO5
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50228 complex | | Descriptor: | MAGNESIUM ION, N-[(2S)-1-(hydroxyamino)-3-methyl-3-{[(oxetan-3-yl)methyl]sulfonyl}-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOO
 
 | | Co-Crystal structure of P. aeruginosa LpxC-achn975 complex | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MO4
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50067 complex | | Descriptor: | MAGNESIUM ION, N-[(2R)-1-(hydroxyamino)-3-methyl-3-(methylsulfonyl)-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOD
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50432 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-[(1S)-2-(hydroxyamino)-1-(3-methoxy-1,1-dioxo-1lambda~6~-thietan-3-yl)-2-oxoethyl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, ... | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
3WRU
 
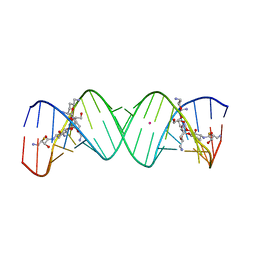 | | Crystal structure of the bacterial ribosomal decoding site in complex with synthetic aminoglycoside with F-HABA group | | Descriptor: | (2R,3R)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranosyl)oxy]-3-{[3-O-(2,6-diamino-2,3,4,6-tetradeoxy-beta-L-threo-hexopyranosyl)-beta-D-ribofuranosyl]oxy}-2-hydroxycyclohexyl]-3-fluoro-2-hydroxybutanamide, POTASSIUM ION, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J, Hanessian, S. | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Toxicity Modulation, Resistance Enzyme Evasion, and A-Site X-ray Structure of Broad-Spectrum Antibacterial Neomycin Analogs
Acs Chem.Biol., 9, 2014
|
|
