3T0Z
 
 | | Hsp90 N-terminal domain bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
6CYZ
 
 | |
6JLE
 
 | | Crystal structure of MORN4/Myo3a complex | | Descriptor: | CITRIC ACID, GLYCEROL, MORN repeat-containing protein 4, ... | | Authors: | Li, J, Liu, H, Raval, M.H, Wan, J, Yengo, C.M, Liu, W, Zhang, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the MORN4/Myo3a Tail Complex Reveals MORN Repeats as Protein Binding Modules.
Structure, 27, 2019
|
|
3FP2
 
 | | Crystal structure of Tom71 complexed with Hsp82 C-terminal fragment | | Descriptor: | ATP-dependent molecular chaperone HSP82, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP3
 
 | | Crystal structure of Tom71 | | Descriptor: | CHLORIDE ION, SULFATE ION, TPR repeat-containing protein YHR117W | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP4
 
 | | Crystal structure of Tom71 complexed with Ssa1 C-terminal fragment | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
2O9Q
 
 | | The crystal structure of Bovine Trypsin complexed with a small inhibition peptide ORB2K | | Descriptor: | CALCIUM ION, Cationic trypsin, ORB2K, ... | | Authors: | Li, J, Zhang, C, Xu, X, Wang, J, Gong, W, Lai, R. | | Deposit date: | 2006-12-14 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From protease inhibitor to antibiotics: single point mutation makes tremendous functional shift
To be Published
|
|
8EW5
 
 | | The structure of flightin within myosin thick filaments from Bombus ignitus flight muscle | | Descriptor: | Flightin | | Authors: | Li, J, Rahmani, H, Abbasi Yeganeh, F, Rastegarpouyani, H, Taylor, D.W, Wood, N.B, Previs, M.J, Iwamoto, H, Taylor, K.A. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-04 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Flight Muscle Thick Filament from the Bumble Bee, Bombus ignitus , at 6 angstrom Resolution.
Int J Mol Sci, 24, 2022
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8W6I
 
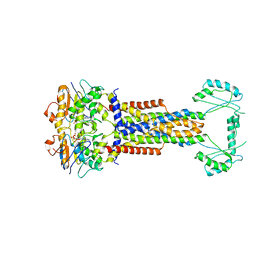 | |
7U20
 
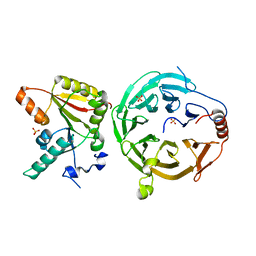 | | Crystal structure of human METTL1 and WDR4 complex | | Descriptor: | SULFATE ION, tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Li, J, Nowak, R.P, Fischer, E.S, Gregory, R. | | Deposit date: | 2022-02-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
8W6J
 
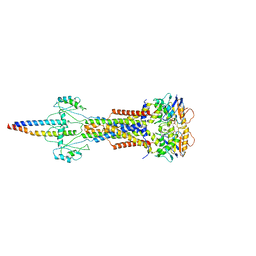 | | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, He, Y, Luo, M. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc
To Be Published
|
|
3RY7
 
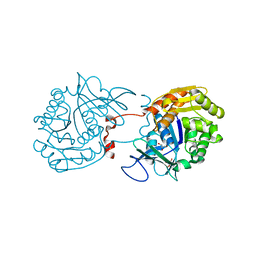 | | Crystal Structure of Sa239 | | Descriptor: | GLYCEROL, Ribokinase | | Authors: | Li, J, Wu, M, Wang, L, Zang, J. | | Deposit date: | 2011-05-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Sa239 reveals the structural basis for the activation of ribokinase by monovalent cations.
J.Struct.Biol., 177, 2012
|
|
4O7P
 
 | | Crystal structure of Mycobacterium tuberculosis maltose kinase MaK complexed with maltose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Maltokinase, SULFATE ION, ... | | Authors: | Li, J, Guan, X.T, Rao, Z.H. | | Deposit date: | 2013-12-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Homotypic dimerization of a maltose kinase for molecular scaffolding.
Sci Rep, 4, 2014
|
|
4O7O
 
 | |
7XC3
 
 | |
7XC4
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain 3 (SARS-unique domain-M) in complex with Oxaprozin | | Descriptor: | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid, Papain-like protease nsp3 | | Authors: | Li, J, Liu, Y, Gao, J, Ruan, K. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Binding Sites of SARS-CoV-2 Macrodomain 3 Probed by Oxaprozin and Meclomen.
J.Med.Chem., 65, 2022
|
|
7BV3
 
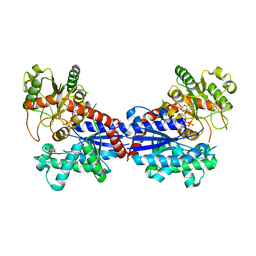 | | Crystal structure of a ugt transferase from Siraitia grosvenorii in complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases
Green Synth Catal, 2021
|
|
4L93
 
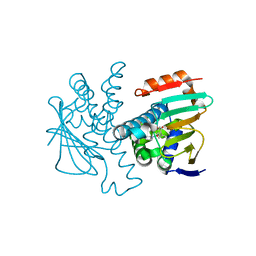 | | Crystal structure of Human Hsp90 with S36 | | Descriptor: | 3,4-dihydroisoquinolin-2(1H)-yl[2,4-dihydroxy-5-(propan-2-yl)phenyl]methanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Crystal structure of Human Hsp90 with S36
To be Published
|
|
4L90
 
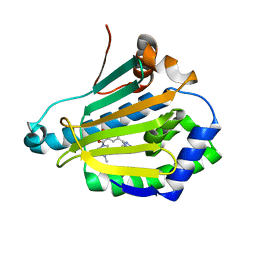 | | Crystal structure of Human Hsp90 with RL3 | | Descriptor: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](4-methylpiperazin-1-yl)methanone | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Human Hsp90 with RL3
to be published
|
|
2A7R
 
 | | Crystal structure of human Guanosine Monophosphate reductase 2 (GMPR2) | | Descriptor: | GMP reductase 2, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Li, J, Wei, Z, Zheng, M, Gu, X, Deng, Y, Qiu, R, Chen, F, Ji, C, Gong, W, Xie, Y, Mao, Y. | | Deposit date: | 2005-07-05 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Guanosine Monophosphate Reductase 2 (GMPR2) in Complex with GMP
J.Mol.Biol., 355, 2006
|
|
2AD7
 
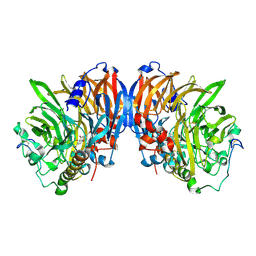 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) in the presence of methanol | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
2AD6
 
 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
2AD8
 
 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) in the presence of ethanol | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
