6RWO
 
 | | SIVrcm intasome (Q148H/G140S) in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
5ZNZ
 
 | | Structure of mDR3 DD with MBP tag mutant-I387V | | Descriptor: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | Authors: | Jin, T, Yin, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6RWL
 
 | | SIVrcm intasome | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*AP*AP*GP*AP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*CP*CP*A)-3'), Pol protein, ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWN
 
 | | SIVrcm intasome in complex with dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWM
 
 | | SIVrcm intasome in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6JEH
 
 | |
6JEG
 
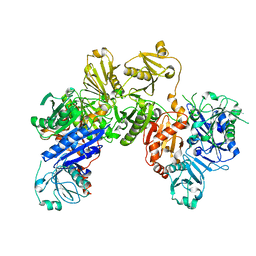 | |
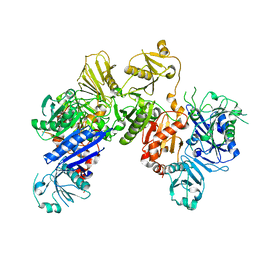
6JCO
 
 | |
6SKM
 
 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,12) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-26 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLQ
 
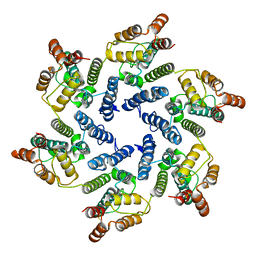 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-12,11) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLU
 
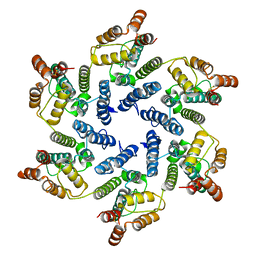 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,11) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6CN5
 
 | |
6CN6
 
 | | RORC2 LBD complexed with compound 34 | | Descriptor: | 3-cyano-N-{3-[1-(cyclopentanecarbonyl)piperidin-4-yl]-1,4-dimethyl-1H-indol-5-yl}benzamide, Nuclear receptor ROR-gamma | | Authors: | Kauppi, B, Vajdos, F. | | Deposit date: | 2018-03-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of 3-Cyano- N-(3-(1-isobutyrylpiperidin-4-yl)-1-methyl-4-(trifluoromethyl)-1 H-pyrrolo[2,3- b]pyridin-5-yl)benzamide: A Potent, Selective, and Orally Bioavailable Retinoic Acid Receptor-Related Orphan Receptor C2 Inverse Agonist.
J. Med. Chem., 61, 2018
|
|
6DJ4
 
 | |
6DAQ
 
 | | PhdJ bound to substrate intermediate | | Descriptor: | PhdJ | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-10 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
6DAN
 
 | | PhdJ WT 2 Angstroms resolution | | Descriptor: | CHLORIDE ION, PhdJ | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
6DAO
 
 | | NahE WT selenomethionine | | Descriptor: | Trans-O-hydroxybenzylidenepyruvate hydratase-aldolase | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
4OE1
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 (C256S/C430S/C449S) in complex with an 18-nt PSAJ rna element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Li, Q, Yan, C, Wu, J, Yin, P, Yan, N. | | Deposit date: | 2014-01-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Examination of the dimerization states of the single-stranded RNA recognition protein pentatricopeptide repeat 10 (PPR10).
J.Biol.Chem., 289, 2014
|
|
6E4Z
 
 | |
6E4Y
 
 | |
7XPQ
 
 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPP
 
 | |
