1Z2F
 
 | | solution structure of CfAFP-501 | | Descriptor: | Antifreeze Protein Isoform 501 | | Authors: | Li, C, Jin, C. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Antifreeze Protein CfAFP-501 from Choristoneura fumiferana
J.Biomol.Nmr, 32, 2005
|
|
8YKW
 
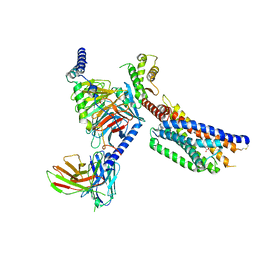 | | Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8YKV
 
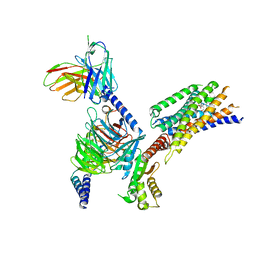 | | Cryo-EM structure of succinate receptor SUCR1 bound to compound 31 | | Descriptor: | (2~{S})-2-[[6-[4-(trifluoromethyloxy)phenyl]pyridin-2-yl]carbonylamino]butanedioic acid, Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8YKX
 
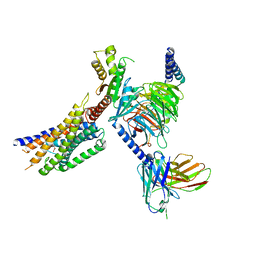 | | Cryo-EM structure of succinate receptor SUCR1 bound to maleic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
5YEI
 
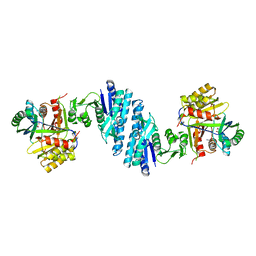 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
7QOX
 
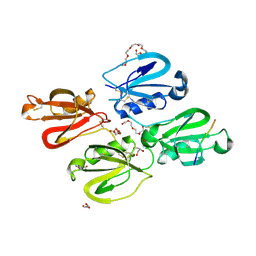 | | Factor XI and Plasma Kallikrein apple domain structures reveals different kininogen bound complexes | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Li, C, Awital, B, Wong, S, Dreveny, I, Meijers, J, Emsley, J. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J Thromb Haemost, 17, 2019
|
|
7QOT
 
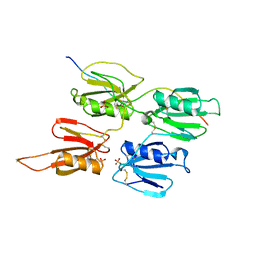 | | Factor XI and Plasma Kallikrein apple domain structures reveals different kininogen bound complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa heavy chain, Kininogen-1 light chain, ... | | Authors: | Li, C, Awital, B, Wong, S, Dreveny, I, Meijers, J, Emsley, J. | | Deposit date: | 2021-12-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J Thromb Haemost, 17, 2019
|
|
8QR1
 
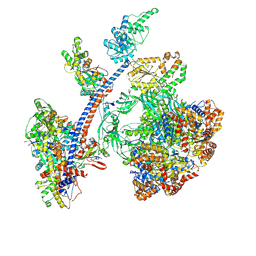 | | Cryo-EM structure of the human Tip60 complex | | Descriptor: | Actin, cytoplasmic 1, N-terminally processed, ... | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 635, 2024
|
|
8QRI
 
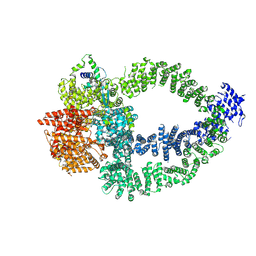 | | TRRAP and EP400 in the human Tip60 complex | | Descriptor: | E1A-binding protein p400, Transformation/transcription domain-associated protein | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, SChultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 635, 2024
|
|
8Y51
 
 | | Cryo-EM structure of the BRS3-Gq complex | | Descriptor: | Bombesin receptor subtype-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Xu, Y, Yin, W, Xu, H.E. | | Deposit date: | 2024-01-31 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of self-activation and ligand recognition of human bombesin receptor subtype-3
To Be Published
|
|
2XV3
 
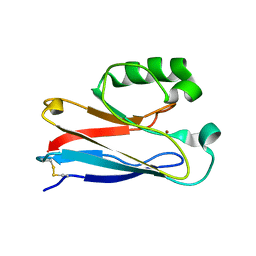 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAAAHAAAAM), chemically reduced, pH5.3 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
2XV0
 
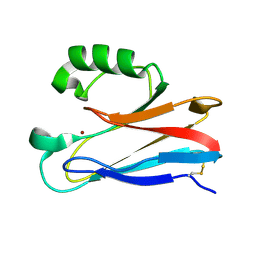 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.8 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
2XV2
 
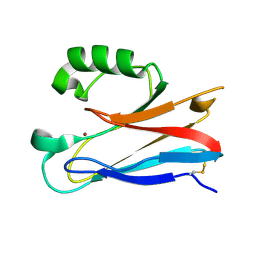 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.2 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
1KZZ
 
 | | DOWNSTREAM REGULATOR TANK BINDS TO THE CD40 RECOGNITION SITE ON TRAF3 | | Descriptor: | TNF receptor associated factor 3, TRAF family member-associated NF-kappa-b activator | | Authors: | Li, C, Ni, C.-Z, Havert, M.L, Cabezas, E, He, J, Kaiser, D, Reed, J.C, Satterthwait, A.C, Cheng, G, Ely, K.R. | | Deposit date: | 2002-02-08 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Downstream regulator TANK binds to the CD40 recognition site on TRAF3.
Structure, 10, 2002
|
|
3ERC
 
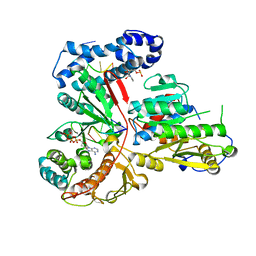 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase with three fragments of RNA and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
3ER9
 
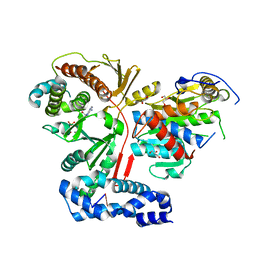 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with UU and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-R(UP*U)-3', CALCIUM ION, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
3ER8
 
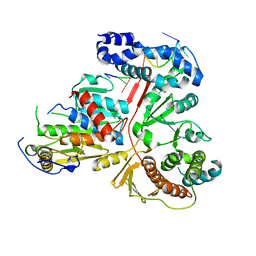 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with two fragments of RNA | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit, RNA/DNA chimera 5'-D(CP*CP*)R(UP*UP*)D(C)-3', ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
8RRS
 
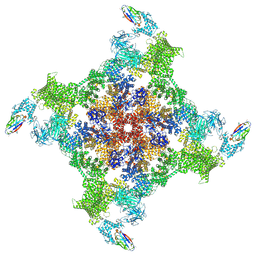 | | Structure of mouse RyR2 solubilised in detergent in open state in complex with Ca2+, ATP, caffeine and Nb9657. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
8RRW
 
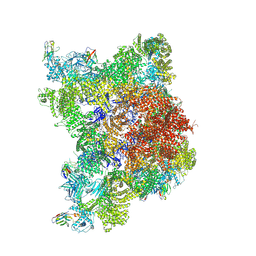 | |
8RRV
 
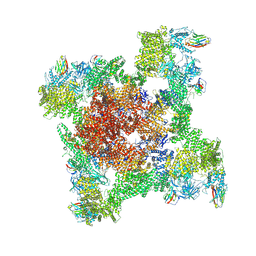 | |
8RRT
 
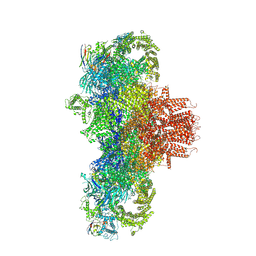 | |
8RRX
 
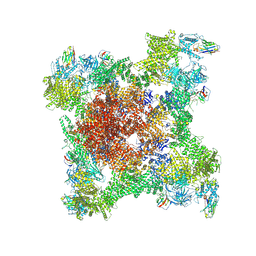 | | Structure of RyR1 reconstituted into lipid nanodisc in primed state in complex with Ca2+, ATP, caffeine and Nb9657 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
8RS0
 
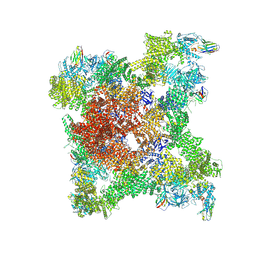 | |
8RRU
 
 | |
3QDP
 
 | |
